|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DEF-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
200 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
1.82 |
-16.65 |
2 |
5 |
0 |
68 |
245.662 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.16 |
3.04 |
-55.01 |
1 |
5 |
-1 |
71 |
244.654 |
4 |
↓
|
|
|
|
Analogs
-
3871393
-
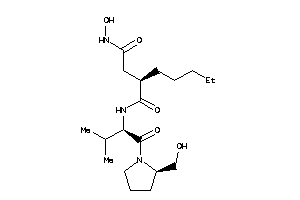
-
3871392
-
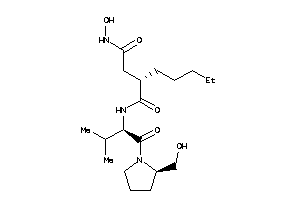
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DEF-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
5 |
0.43 |
Binding ≤ 10μM
|
|
Q9JN24-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
140 |
0.36 |
Binding ≤ 10μM |
|
DEF1A-1-E |
Peptide Deformylase 1A, Chloroplastic (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.39 |
Binding ≤ 10μM
|
|
DEFM-1-E |
Peptide Deformylase Mitochondrial (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.32 |
Binding ≤ 10μM
|
|
ECE1-1-E |
Endothelin-converting Enzyme 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.28 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.34 |
Binding ≤ 10μM |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.31 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.27 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
190 |
0.35 |
Binding ≤ 10μM |
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
330 |
0.34 |
Binding ≤ 10μM |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.26 |
Binding ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
2512 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
2.32 |
-16.44 |
4 |
8 |
0 |
119 |
385.505 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.51 |
3.4 |
-55.95 |
3 |
8 |
-1 |
122 |
384.497 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DEF-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
330 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.72 |
1.1 |
-12.45 |
4 |
5 |
0 |
84 |
237.284 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DEF-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
330 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.72 |
1.1 |
-12.45 |
4 |
5 |
0 |
84 |
237.284 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DEF-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
5 |
0.73 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.41 |
1.19 |
-15.69 |
3 |
5 |
0 |
78 |
238.268 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.41 |
2.4 |
-55.94 |
2 |
5 |
-1 |
81 |
237.26 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DEF-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
5 |
0.73 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.41 |
1.19 |
-15.65 |
3 |
5 |
0 |
78 |
238.268 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.41 |
2.4 |
-55.94 |
2 |
5 |
-1 |
81 |
237.26 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DEF-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
74 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.69 |
3.37 |
-16.91 |
2 |
5 |
0 |
67 |
252.295 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DEF-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
74 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.69 |
3.37 |
-16.88 |
2 |
5 |
0 |
67 |
252.295 |
3 |
↓
|
|