|
|
Analogs
-
30691414
-
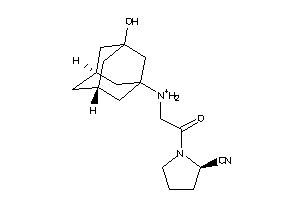
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.51 |
Binding ≤ 10μM
|
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.32 |
Binding ≤ 10μM |
|
DPP9-1-E |
Dipeptidyl Peptidase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
680 |
0.39 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
4.11 |
-49.86 |
3 |
5 |
1 |
81 |
304.414 |
3 |
↓
|
|
|
|
Analogs
-
37823165
-
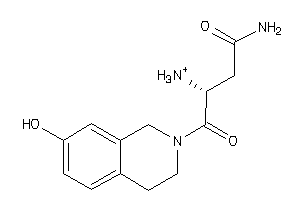
-
37823166
-
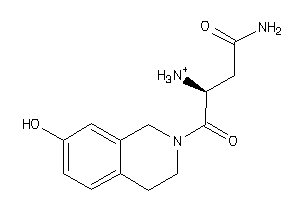
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
555 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
7.57 |
-43.01 |
3 |
5 |
1 |
68 |
350.442 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.30 |
Binding ≤ 10μM
|
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.30 |
Binding ≤ 10μM |
|
DPP9-1-E |
Dipeptidyl Peptidase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.28 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
10.86 |
-47.58 |
3 |
6 |
1 |
71 |
505.589 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.75 |
10.53 |
-13.77 |
2 |
6 |
0 |
70 |
504.581 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.27 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
10.98 |
-46.66 |
3 |
6 |
1 |
71 |
519.616 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.08 |
10.66 |
-13.73 |
2 |
6 |
0 |
70 |
518.608 |
6 |
↓
|
|
|
|
Analogs
-
3931585
-
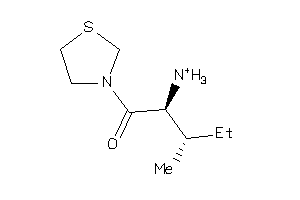
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.68 |
Binding ≤ 10μM
|
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.62 |
Binding ≤ 10μM
|
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.60 |
Binding ≤ 10μM
|
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.72 |
Binding ≤ 10μM
|
|
DPP9-1-E |
Dipeptidyl Peptidase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.53 |
2.53 |
-44.92 |
3 |
3 |
1 |
48 |
203.331 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.74 |
Binding ≤ 10μM |
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.67 |
Binding ≤ 10μM
|
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.65 |
Binding ≤ 10μM
|
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
27 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.99 |
4.47 |
-52.97 |
3 |
4 |
1 |
72 |
236.339 |
2 |
↓
|
|
|
|
Analogs
-
37823165
-

-
37823166
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
6.77 |
-48.81 |
3 |
7 |
1 |
87 |
410.494 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3016 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
6.99 |
-45.49 |
3 |
3 |
1 |
48 |
273.4 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
3203426
-
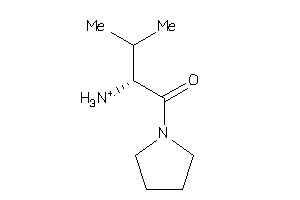
-
35931795
-
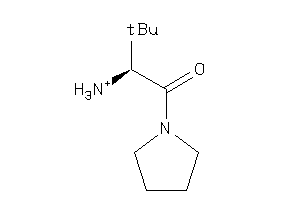
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
470 |
0.74 |
Binding ≤ 10μM |
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5400 |
0.61 |
Binding ≤ 10μM
|
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5200 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.05 |
2.43 |
-41.47 |
3 |
3 |
1 |
48 |
171.264 |
2 |
↓
|
|
|
|
Analogs
-
3964523
-
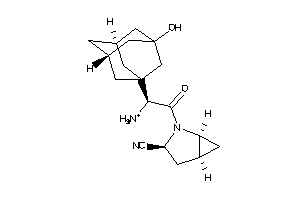
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
240 |
0.40 |
Binding ≤ 10μM
|
|
DPP9-1-E |
Dipeptidyl Peptidase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
71 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
2.91 |
-12.9 |
3 |
5 |
0 |
90 |
315.417 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.56 |
Binding ≤ 10μM
|
|
DPP9-1-E |
Dipeptidyl Peptidase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
5.07 |
-46.45 |
3 |
3 |
1 |
48 |
233.335 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.14 |
4.78 |
-6.12 |
2 |
3 |
0 |
46 |
232.327 |
3 |
↓
|
|
|
|
Analogs
-
37823101
-
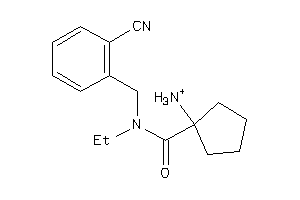
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
6.24 |
-46.52 |
3 |
3 |
1 |
48 |
259.373 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1320 |
0.48 |
Binding ≤ 10μM
|
|
DPP9-1-E |
Dipeptidyl Peptidase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
5.08 |
-45.44 |
3 |
3 |
1 |
48 |
233.335 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.14 |
4.84 |
-6.73 |
2 |
3 |
0 |
46 |
232.327 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
147 |
0.35 |
Binding ≤ 10μM
|
|
DPP9-1-E |
Dipeptidyl Peptidase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
168 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
-4.15 |
-55.68 |
4 |
9 |
1 |
132 |
379.437 |
10 |
↓
|
|
|
|
Analogs
-
13522765
-
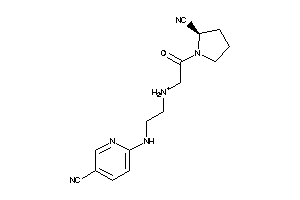
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP2-2-E |
Dipeptidyl Peptidase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6600 |
0.33 |
Binding ≤ 10μM
|
|
DPP4-1-E |
Dipeptidyl Peptidase IV (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4573 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.38 |
4.95 |
-65.32 |
3 |
7 |
1 |
109 |
299.358 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.38 |
3.53 |
-18.36 |
2 |
7 |
0 |
105 |
298.35 |
6 |
↓
|
|
|
|
|