|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.22 |
Binding ≤ 10μM |
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.24 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
14.5 |
-48.94 |
0 |
8 |
-1 |
110 |
511.963 |
8 |
↓
|
|
|
|
Analogs
-
8425894
-
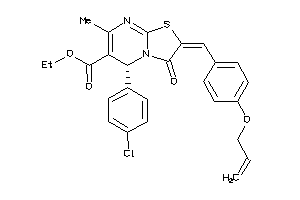
-
8425895
-
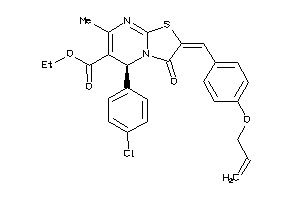
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.22 |
Binding ≤ 10μM |
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.24 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
14.48 |
-48.78 |
0 |
8 |
-1 |
110 |
511.963 |
8 |
↓
|
|
|
|
Analogs
-
871881
-
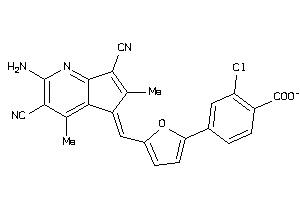
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.24 |
Binding ≤ 10μM
|
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3870 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
10.84 |
-61.32 |
2 |
7 |
-1 |
140 |
441.854 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.22 |
Binding ≤ 10μM
|
|
PP1A-2-E |
Serine/threonine Protein Phosphatase PP1-alpha Catalytic Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8100 |
0.21 |
Binding ≤ 10μM
|
|
PP1G-2-E |
Serine/threonine Protein Phosphatase PP1-gamma Catalytic Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8100 |
0.21 |
Binding ≤ 10μM
|
|
PPAC-1-E |
Low Molecular Weight Phosphotyrosine Protein Phosphatase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.23 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4800 |
0.22 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9900 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.75 |
0.27 |
-98.84 |
3 |
12 |
-2 |
188 |
510.55 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.75 |
0.34 |
-185.53 |
2 |
12 |
-3 |
190 |
509.542 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.75 |
0.34 |
-189.06 |
2 |
12 |
-3 |
190 |
509.542 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.30 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
640 |
0.36 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
3.23 |
-41.73 |
1 |
5 |
-1 |
86 |
451.094 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.25 |
3.99 |
-92.12 |
0 |
5 |
-2 |
88 |
450.086 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.28 |
5.32 |
-46.11 |
0 |
5 |
-1 |
82 |
451.094 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Binding ≤ 10μM
|
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.48 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
95 |
0.45 |
Binding ≤ 10μM
|
|
MPIP3-2-E |
Dual Specificity Phosphatase Cdc25C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
89 |
0.45 |
Binding ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
2300 |
0.36 |
Functional ≤ 10μM
|
|
Z80800-1-O |
MIA PaCa-2 (Pancreatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2500 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.42 |
1.28 |
-9.59 |
0 |
6 |
0 |
72 |
321.764 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.42 |
1.17 |
-10.76 |
0 |
6 |
0 |
72 |
321.764 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.42 |
3.55 |
-37.71 |
1 |
6 |
1 |
73 |
322.772 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2090 |
0.44 |
Binding ≤ 10μM
|
|
LEF-1-B |
Anthrax Lethal Factor (cluster #1 Of 2), Bacterial |
Bacteria |
4600 |
0.42 |
Binding ≤ 10μM
|
|
Q9AQH7-1-B |
DnaC Helicase (cluster #1 Of 1), Bacterial |
Bacteria |
4000 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
9.44 |
-43.47 |
0 |
3 |
-1 |
45 |
260.338 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1760 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.09 |
11.7 |
-21.94 |
1 |
8 |
0 |
114 |
451.438 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-1-E |
Dual Specificity Protein Phosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.19 |
Binding ≤ 10μM
|
|
DUS3-2-E |
Dual Specificity Protein Phosphatase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.23 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2200 |
0.20 |
Binding ≤ 10μM
|
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.20 |
Binding ≤ 10μM
|
|
PTPRC-3-E |
Leukocyte Common Antigen (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1800 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.52 |
8.73 |
-53.3 |
0 |
10 |
-1 |
134 |
617.776 |
8 |
↓
|
|
|
|
Analogs
-
2395351
-
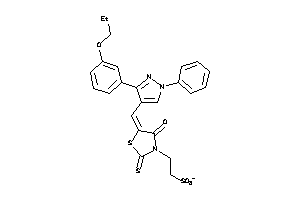
-
19878963
-
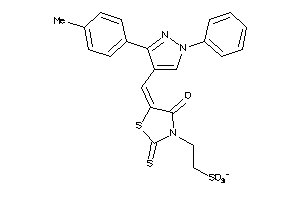
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-2-E |
Dual Specificity Protein Phosphatase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3100 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
9.44 |
-50.62 |
0 |
7 |
-1 |
97 |
484.604 |
6 |
↓
|
|
|
|
Analogs
-
6645245
-
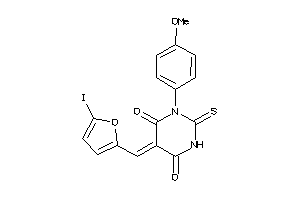
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4440 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
-0.74 |
-11.83 |
1 |
6 |
0 |
77 |
454.245 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-1-E |
Dual Specificity Protein Phosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.22 |
Binding ≤ 10μM
|
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.27 |
Binding ≤ 10μM
|
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.20 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
460 |
0.22 |
Binding ≤ 10μM
|
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
620 |
0.22 |
Binding ≤ 10μM
|
|
PTPRC-1-E |
Leukocyte Common Antigen (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
12.53 |
-51.54 |
0 |
8 |
-1 |
106 |
611.146 |
9 |
↓
|
|
|
|
Analogs
-
35517797
-

-
1452085
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1860 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.96 |
7.53 |
-19.75 |
1 |
6 |
0 |
92 |
332.362 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-1-E |
Dual Specificity Protein Phosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
780 |
0.22 |
Binding ≤ 10μM
|
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
78 |
0.26 |
Binding ≤ 10μM
|
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.20 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1800 |
0.21 |
Binding ≤ 10μM
|
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.21 |
Binding ≤ 10μM
|
|
PTPRC-1-E |
Leukocyte Common Antigen (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
610 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
12.04 |
-51.46 |
0 |
8 |
-1 |
106 |
576.701 |
9 |
↓
|
|
|
|
Analogs
-
441051
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3490 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
6.85 |
-44.38 |
1 |
5 |
-1 |
74 |
330.776 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2940 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.18 |
-0.62 |
-18.47 |
2 |
3 |
0 |
37 |
368.461 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-1-E |
Dual Specificity Protein Phosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
470 |
0.22 |
Binding ≤ 10μM
|
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
71 |
0.25 |
Binding ≤ 10μM
|
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.19 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
590 |
0.22 |
Binding ≤ 10μM
|
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.21 |
Binding ≤ 10μM
|
|
PTPRC-1-E |
Leukocyte Common Antigen (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
12.62 |
-50.63 |
0 |
8 |
-1 |
106 |
611.146 |
9 |
↓
|
|
|
|
Analogs
-
19912604
-

-
2395351
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1800 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.36 |
-6.55 |
-51.07 |
0 |
8 |
-1 |
106 |
500.603 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5100 |
0.19 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.24 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8700 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.48 |
3.16 |
-41.83 |
4 |
12 |
-1 |
190 |
519.49 |
7 |
↓
|
|
|
|
Analogs
-
5339636
-
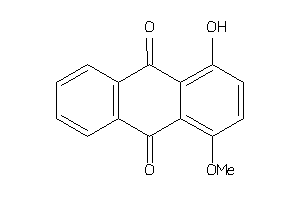
-
6127508
-
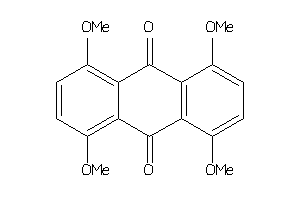
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-3-E |
Dual Specificity Protein Phosphatase 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3070 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
8.11 |
-14.1 |
0 |
4 |
0 |
53 |
268.268 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2640 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
5.84 |
-8.31 |
1 |
7 |
0 |
94 |
364.155 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.91 |
5.7 |
-32.6 |
0 |
7 |
-1 |
92 |
363.147 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.91 |
5.68 |
-32.73 |
0 |
7 |
-1 |
92 |
363.147 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-1-E |
Dual Specificity Protein Phosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.22 |
Binding ≤ 10μM
|
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
74 |
0.25 |
Binding ≤ 10μM
|
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.19 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
420 |
0.22 |
Binding ≤ 10μM
|
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
870 |
0.21 |
Binding ≤ 10μM
|
|
PTPRC-1-E |
Leukocyte Common Antigen (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
12.21 |
-50.97 |
0 |
8 |
-1 |
106 |
594.691 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-2-E |
Dual Specificity Protein Phosphatase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2400 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.54 |
6.28 |
-54.68 |
0 |
11 |
-1 |
144 |
619.748 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-2-E |
Dual Specificity Protein Phosphatase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1800 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
9.3 |
-50.14 |
0 |
7 |
-1 |
97 |
505.022 |
6 |
↓
|
|
|
|
Analogs
-
2967486
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3130 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.81 |
-7.13 |
-52.2 |
0 |
6 |
-1 |
92 |
428.92 |
5 |
↓
|
|
|
|
Analogs
-
4077011
-
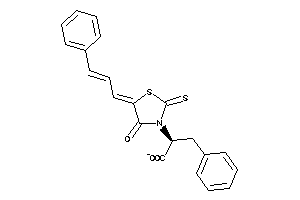
-
33861928
-
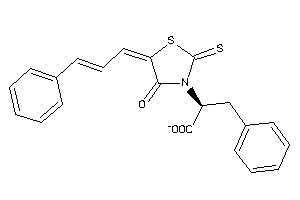
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-3-E |
Dual Specificity Protein Phosphatase 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2010 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
0.11 |
-60.35 |
0 |
4 |
-1 |
62 |
394.497 |
6 |
↓
|
|
|
|
Analogs
-
33861928
-

-
1269245
-
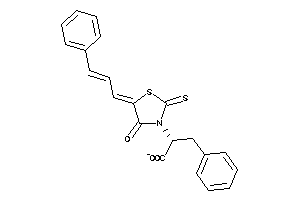
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-3-E |
Dual Specificity Protein Phosphatase 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2010 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
0.12 |
-60.38 |
0 |
4 |
-1 |
62 |
394.497 |
6 |
↓
|
|