|
|
Analogs
-
39260312
-
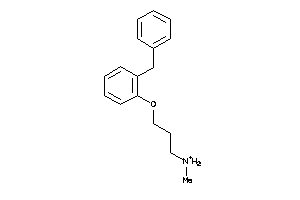
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-4-E |
Squalene Synthetase (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
12 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.64 |
10.75 |
-40.57 |
2 |
2 |
1 |
26 |
284.423 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-4-E |
Squalene Synthetase (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
56 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
8.25 |
-38.48 |
2 |
2 |
1 |
26 |
234.363 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-3-E |
Squalene Synthetase (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2300 |
0.39 |
Binding ≤ 10μM |
|
FNTA-2-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
FNTB-2-E |
Protein Farnesyltransferase Beta Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.76 |
4.93 |
-45.96 |
2 |
4 |
-1 |
81 |
301.343 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-3-E |
Squalene Synthetase (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2300 |
0.39 |
Binding ≤ 10μM |
|
FNTA-2-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
FNTB-2-E |
Protein Farnesyltransferase Beta Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.76 |
4.94 |
-47.26 |
2 |
4 |
-1 |
81 |
301.343 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
86599
-
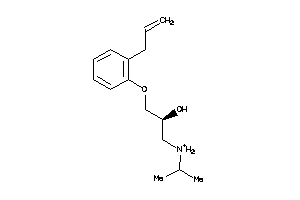
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
5.16 |
-40.25 |
3 |
3 |
1 |
46 |
250.362 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
3.88 |
-5.22 |
2 |
3 |
0 |
41 |
249.354 |
8 |
↓
|
|
|
|
Analogs
-
3647816
-
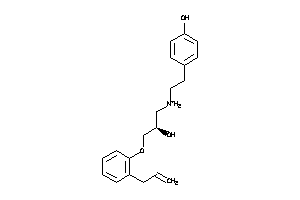
-
3647817
-
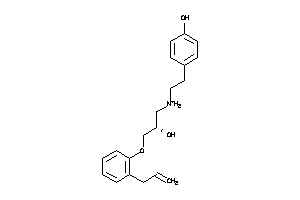
-
23
-
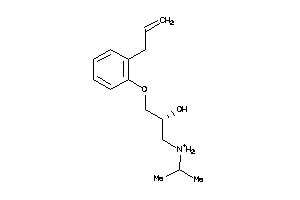
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
5.19 |
-40.14 |
3 |
3 |
1 |
46 |
250.362 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
4.01 |
-5.14 |
2 |
3 |
0 |
41 |
249.354 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-3-E |
Squalene Synthetase (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
10.16 |
-38.13 |
2 |
2 |
1 |
16 |
293.434 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-3-E |
Squalene Synthetase (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
10.24 |
-35.54 |
2 |
2 |
1 |
16 |
293.434 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.44 |
4.21 |
-123.14 |
1 |
7 |
-2 |
127 |
384.346 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
14242978
-
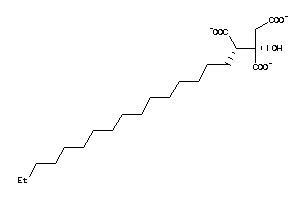
-
14242979
-
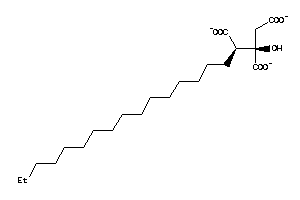
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.24 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.12 |
14.8 |
-228.05 |
1 |
7 |
-3 |
141 |
413.531 |
20 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.12 |
11.29 |
-57.33 |
3 |
7 |
-1 |
135 |
415.547 |
20 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.12 |
13.27 |
-125.06 |
2 |
7 |
-2 |
138 |
414.539 |
20 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 33 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-2-E |
Squalene Synthetase (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
640 |
0.46 |
Binding ≤ 10μM
|
|
FPPS-1-E |
Farnesyl Diphosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
480 |
0.47 |
Binding ≤ 10μM
|
|
Z50725-4-O |
Trypanosoma Brucei Rhodesiense (cluster #4 Of 7), Other |
Other |
960 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.56 |
-0.37 |
-173.95 |
3 |
8 |
-2 |
148 |
317.215 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.56 |
0.81 |
-304.39 |
2 |
8 |
-3 |
151 |
316.207 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.56 |
-1.47 |
-87.19 |
4 |
8 |
-1 |
145 |
318.223 |
9 |
↓
|
|