|
|
|
|
|
|
|
|
Analogs
-
33854099
-
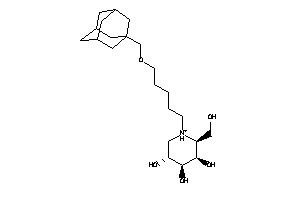
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CEGT-1-E |
Ceramide Glucosyltransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
150 |
0.34 |
Binding ≤ 10μM
|
|
GBA2-1-E |
Beta-glucosidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.38 |
Binding ≤ 10μM |
|
GLCM-2-E |
Glucosylceramidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.28 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
2.91 |
-31.36 |
5 |
6 |
1 |
95 |
398.564 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.81 |
3.44 |
-31.92 |
4 |
6 |
0 |
97 |
397.556 |
9 |
↓
|
|
|
|
|
|
|
Analogs
-
22053713
-
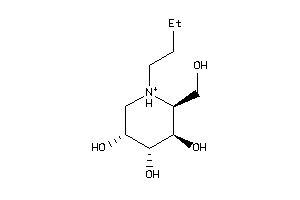
-
3957104
-
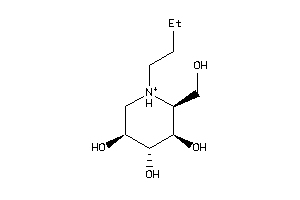
-
2562349
-
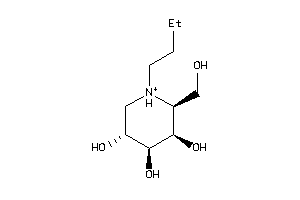
-
3794711
-
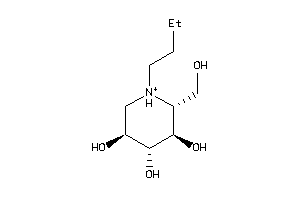
Draw
Identity
99%
90%
80%
70%
Popular Name:
3,4,5-Piperidinetriol, 1-butyl-2-(hydroxymethyl)-, (2S,3S,4R,5S)- (9CI)
3,4,5-Piperidinetriol, 1-butyl-2…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBA2-1-E |
Beta-glucosidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.72 |
-4.84 |
-32.86 |
5 |
5 |
1 |
85 |
220.289 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CEGT-1-E |
Ceramide Glucosyltransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM |
|
GBA2-1-E |
Beta-glucosidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.56 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
0.83 |
-29.77 |
5 |
5 |
1 |
85 |
290.424 |
9 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBA2-1-E |
Beta-glucosidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
1.6 |
-34.72 |
5 |
5 |
1 |
85 |
332.505 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.32 |
2.06 |
-42.35 |
4 |
5 |
0 |
88 |
331.497 |
12 |
↓
|
|
|
|
|
|
|
|