|
|
|
|
|
Analogs
-
8582318
-
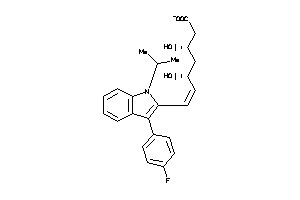
-
8582319
-
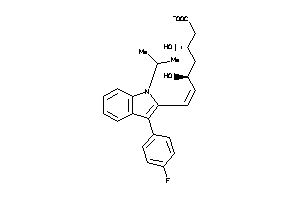
-
1530639
-
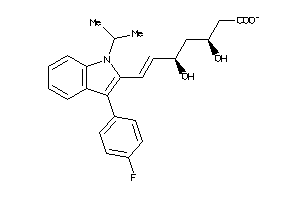
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
11.25 |
-51.38 |
2 |
5 |
-1 |
86 |
410.465 |
8 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
28 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.08 |
10.52 |
-10.79 |
1 |
4 |
0 |
51 |
503.206 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.38 |
Binding ≤ 10μM
|
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
5 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
11.12 |
-14.04 |
1 |
5 |
0 |
73 |
404.547 |
7 |
↓
|
|
|
|
Analogs
-
8101062
-
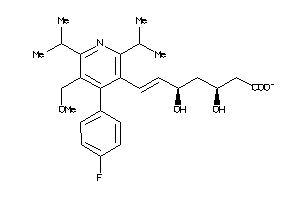
-
9831786
-
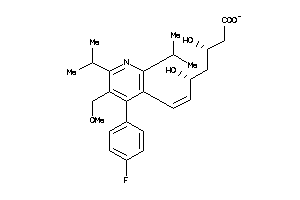
-
9831787
-
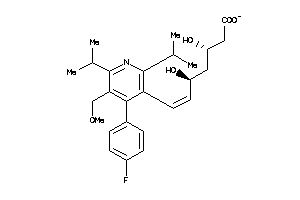
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-2-E |
HMG-CoA Reductase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
Z81135-1-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #1 Of 4), Other |
Other |
7 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
9.79 |
-51.62 |
2 |
6 |
-1 |
103 |
458.55 |
11 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.45 |
10.19 |
-65.02 |
3 |
6 |
0 |
104 |
459.558 |
11 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
43772721
-
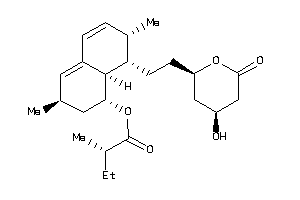
-
43772723
-
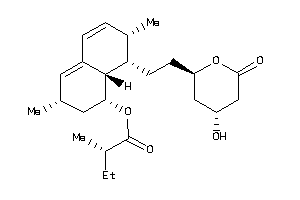
-
43772724
-
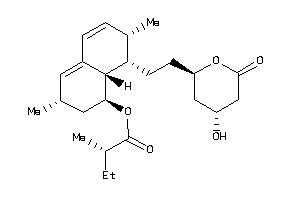
-
43772726
-
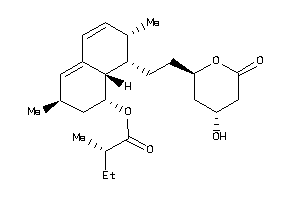
-
43772732
-
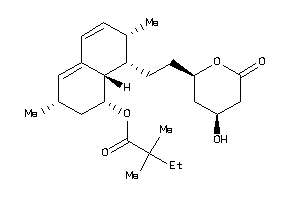
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3R,7S,8S,8aR)-8-[2-[(2S,4R)-4-hydroxy-6-oxo-tetrahydropyran-2-yl]ethyl]-3,7-dimethyl-1,2,3,7,8,
[(1S,3R,7S,8S,8aR)-8-[2-[(2S,4R)…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
11.18 |
-9.38 |
1 |
5 |
0 |
73 |
404.547 |
7 |
↓
|
|
|
|
Analogs
-
43772721
-

-
43772723
-

-
43772724
-

-
43772726
-

-
43772732
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3R,7S,8S,8aS)-8-[2-[(2S,4R)-4-hydroxy-6-oxo-tetrahydropyran-2-yl]ethyl]-3,7-dimethyl-1,2,3,7,8,
[(1S,3R,7S,8S,8aS)-8-[2-[(2S,4R)…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
10.74 |
-11.7 |
1 |
5 |
0 |
73 |
404.547 |
7 |
↓
|
|
|
|
Analogs
-
3780893
-
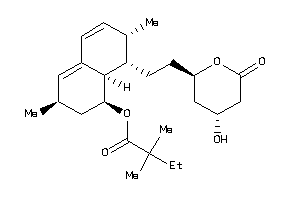
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
2.05 |
-9.95 |
1 |
5 |
0 |
72 |
418.574 |
7 |
↓
|
|
|
|
Analogs
-
3780893
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
11.19 |
-15.42 |
1 |
5 |
0 |
73 |
418.574 |
7 |
↓
|
|
|
|
Analogs
-
3780893
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3R,7S,8S,8aS)-8-[2-[(2S,4R)-4-hydroxy-6-oxo-tetrahydropyran-2-yl]ethyl]-3,7-dimethyl-1,2,3,7,8,
[(1S,3R,7S,8S,8aS)-8-[2-[(2S,4R)…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
11.09 |
-12.03 |
1 |
5 |
0 |
73 |
418.574 |
7 |
↓
|
|
|
|
Analogs
-
3833877
-
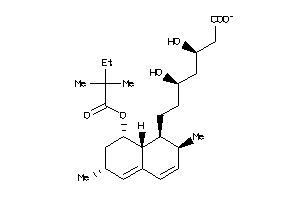
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5R)-7-[(1S,2S,6R,8S,8aR)-2,6-dimethyl-8-[(2R)-2-(trideuteriomethyl)butanoyl]oxy-1,2,6,7,8,8a-hex
(3R,5R)-7-[(1S,2S,6R,8S,8aR)-2,6…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3690 |
0.25 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
10.22 |
-43.55 |
2 |
6 |
-1 |
107 |
421.554 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.15 |
0.91 |
-19.29 |
2 |
6 |
0 |
80 |
540.635 |
8 |
↓
|
|
|
|
|
|
|
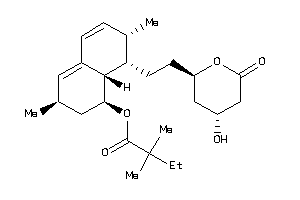
Analogs
-
3984042
-
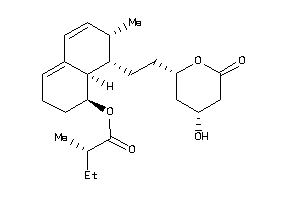
-
4245612
-
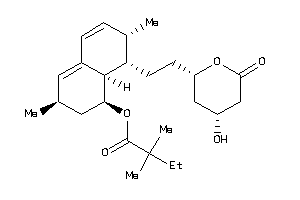
-
4245664
-
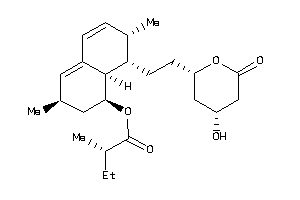
-
4521332
-
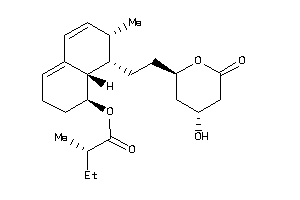
-
26504552
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 51 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.39 |
Binding ≤ 10μM
|
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.39 |
Binding ≤ 10μM
|
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
27 |
0.37 |
Binding ≤ 10μM |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Functional ≤ 10μM
|
|
ICAM1-2-E |
Intercellular Adhesion Molecule-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3780 |
0.26 |
Functional ≤ 10μM
|
|
ITAL-2-E |
Leukocyte Adhesion Glycoprotein LFA-1 Alpha (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3780 |
0.26 |
Functional ≤ 10μM
|
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
25 |
0.37 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
7100 |
0.25 |
Functional ≤ 10μM
|
|
Z80952-1-O |
HES (cluster #1 Of 1), Other |
Other |
17 |
0.38 |
Functional ≤ 10μM
|
|
Z81020-2-O |
HepG2 (Hepatoblastoma Cells) (cluster #2 Of 8), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z81020-2-O |
HepG2 (Hepatoblastoma Cells) (cluster #2 Of 8), Other |
Other |
79 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
11.28 |
-12.84 |
1 |
5 |
0 |
73 |
404.547 |
7 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
10.68 |
-12.93 |
1 |
5 |
0 |
73 |
390.52 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
33681167
-
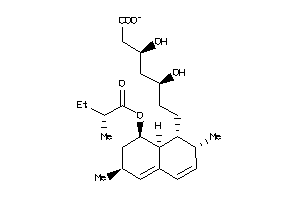
-
33681168
-
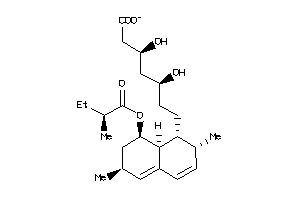
-
33681169
-
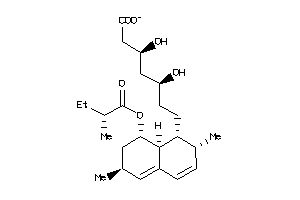
-
4134473
-
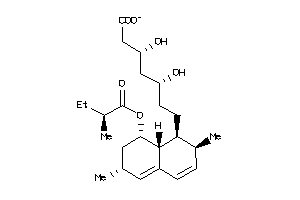
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5S)-7-[(1S,2S,8S,8aR)-2-methyl-8-[(2S)-2-methylbutanoyl]oxy-1,2,6,7,8,8a-hexahydronaphthalen-1-y
(3R,5S)-7-[(1S,2S,8S,8aR)-2-meth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.31 |
8.13 |
-52.9 |
2 |
6 |
-1 |
107 |
407.527 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5R)-7-[(1S,2S,8S,8aS)-2-methyl-8-[(2S)-2-methylbutanoyl]oxy-1,2,6,7,8,8a-hexahydronaphthalen-1-y
(3R,5R)-7-[(1S,2S,8S,8aS)-2-meth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.31 |
9.21 |
-46.29 |
2 |
6 |
-1 |
107 |
407.527 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5S)-7-[(1S,2S,8S,8aS)-2-methyl-8-[(2S)-2-methylbutanoyl]oxy-1,2,6,7,8,8a-hexahydronaphthalen-1-y
(3R,5S)-7-[(1S,2S,8S,8aS)-2-meth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.31 |
7.99 |
-46.39 |
2 |
6 |
-1 |
107 |
407.527 |
11 |
↓
|
|
|
|
Analogs
-
38559082
-
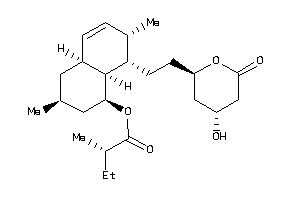
-
5599233
-
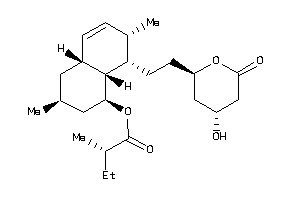
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3S,4aR,7S,8S,8aS)-8-[2-[(2R,4R)-4-hydroxy-6-oxo-tetrahydropyran-2-yl]ethyl]-3,7-dimethyl-1,2,3,
[(1S,3S,4aR,7S,8S,8aS)-8-[2-[(2R…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.36 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
11.35 |
-14.28 |
1 |
5 |
0 |
73 |
406.563 |
7 |
↓
|
|
|
|
Analogs
-
3780893
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
1.46 |
-8.46 |
1 |
5 |
0 |
72 |
404.547 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3R,7S,8S,8aS)-8-[2-[(2S,4R)-4-hydroxy-6-oxo-tetrahydropyran-2-yl]ethyl]-3,7-dimethyl-1,2,3,7,8,
[(1S,3R,7S,8S,8aS)-8-[2-[(2S,4R)…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
10.72 |
-10.63 |
1 |
5 |
0 |
73 |
404.547 |
7 |
↓
|
|