|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3200 |
0.28 |
Binding ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
8.44 |
-16.21 |
2 |
8 |
0 |
97 |
371.441 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.44 |
8.69 |
-42.03 |
3 |
8 |
1 |
99 |
372.449 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.44 |
4.29 |
-37.66 |
3 |
8 |
1 |
99 |
372.449 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
680 |
0.32 |
Binding ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
680 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
3.23 |
-48.36 |
3 |
8 |
-1 |
128 |
367.337 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.54 |
6.13 |
-111.98 |
2 |
8 |
-2 |
130 |
366.329 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6600 |
0.24 |
Binding ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6600 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.01 |
5.92 |
-12.3 |
3 |
5 |
0 |
87 |
419.502 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.01 |
6.08 |
-53.11 |
2 |
5 |
-1 |
89 |
418.494 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
8.76 |
-47 |
4 |
8 |
1 |
105 |
513.385 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.30 |
8.94 |
-2.19 |
0 |
0 |
0 |
0 |
194.343 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.38 |
9.02 |
-99.68 |
5 |
8 |
2 |
106 |
514.393 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6100 |
0.25 |
Binding ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6100 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.91 |
5.23 |
-10.94 |
3 |
5 |
0 |
87 |
470.344 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.91 |
5.28 |
-47.46 |
2 |
5 |
-1 |
89 |
469.336 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.32 |
Binding ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.05 |
3.63 |
-53.27 |
2 |
5 |
-1 |
89 |
396.469 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.05 |
3.59 |
-12.38 |
3 |
5 |
0 |
87 |
397.477 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2200 |
0.27 |
Binding ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.78 |
5.13 |
-10.95 |
3 |
5 |
0 |
87 |
425.893 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.78 |
5.17 |
-47.52 |
2 |
5 |
-1 |
89 |
424.885 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8800 |
0.24 |
Binding ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8800 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.26 |
4.68 |
-11.29 |
3 |
5 |
0 |
87 |
409.438 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.26 |
4.72 |
-47.03 |
2 |
5 |
-1 |
89 |
408.43 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
28 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
2.16 |
-10.37 |
3 |
7 |
0 |
105 |
388.807 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.03 |
2.94 |
-41.12 |
2 |
7 |
-1 |
108 |
387.799 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ENPL-1-E |
Heat Shock Protein 90 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
85 |
0.29 |
Binding ≤ 10μM
|
|
HS90A-1-E |
Heat Shock Protein HSP 90-alpha (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.33 |
Binding ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.33 |
Binding ≤ 10μM
|
|
TRAP1-1-E |
Heat Shock Protein 75 KDa, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
535 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
3.42 |
-10.61 |
3 |
8 |
0 |
108 |
465.55 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.73 |
8.59 |
-100.96 |
3 |
8 |
0 |
112 |
465.55 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.52 |
8.57 |
-101.19 |
3 |
8 |
0 |
112 |
465.55 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-1-E |
Heat Shock Protein HSP 90-alpha (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
775 |
0.21 |
Binding ≤ 10μM
|
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
670 |
0.22 |
Binding ≤ 10μM
|
|
HSP90-1-E |
Heat Shock Protein HSP 90 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.20 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.26 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.25 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.25 |
Functional ≤ 10μM
|
|
HS90A-1-E |
Heat Shock Protein HSP 90-alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.25 |
Functional ≤ 10μM
|
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.25 |
Functional ≤ 10μM
|
|
HSP82-1-F |
Heat Shock Protein HSP 90 (HSP82) (cluster #1 Of 1), Fungal |
Fungi |
4800 |
0.19 |
Binding ≤ 10μM
|
|
Z50347-1-O |
Saccharomyces Cerevisiae (cluster #1 Of 1), Other |
Other |
4800 |
0.19 |
Binding ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
200 |
0.23 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
25 |
0.27 |
Functional ≤ 10μM
|
|
Z80186-4-O |
K562 (Erythroleukemia Cells) (cluster #4 Of 11), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z80224-9-O |
MCF7 (Breast Carcinoma Cells) (cluster #9 Of 14), Other |
Other |
9600 |
0.18 |
Functional ≤ 10μM
|
|
Z80262-1-O |
NCI-H1975 (Bronchoalveolar Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
36 |
0.26 |
Functional ≤ 10μM |
|
Z80359-1-O |
P19 (cluster #1 Of 1), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
70 |
0.25 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
30 |
0.26 |
Functional ≤ 10μM |
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
3 |
0.30 |
Functional ≤ 10μM
|
|
Z81186-1-O |
LS174T (Colon Adencocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
450 |
0.22 |
Functional ≤ 10μM
|
|
Z81331-5-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #5 Of 6), Other |
Other |
6 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
3.25 |
-48.59 |
3 |
11 |
-1 |
170 |
559.636 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
1.28 |
0.74 |
-46.33 |
3 |
11 |
-1 |
171 |
559.636 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.88 |
4.88 |
-17.23 |
3 |
11 |
0 |
164 |
560.644 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
-2.86 |
-11.5 |
2 |
7 |
0 |
88 |
432.303 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
880 |
0.31 |
Binding ≤ 10μM |
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.39 |
Binding ≤ 10μM
|
|
HSC82-1-F |
Heat Shock Protein HSP 90 (HSC82) (cluster #1 Of 1), Fungal |
Fungi |
140 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.68 |
1.48 |
-12.6 |
4 |
7 |
0 |
107 |
387.823 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
3.68 |
1.35 |
-12.68 |
4 |
7 |
0 |
107 |
387.823 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.68 |
2.25 |
-43.73 |
3 |
7 |
-1 |
110 |
386.815 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
0.52 |
-19.74 |
3 |
7 |
0 |
91 |
389.839 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
4.51 |
-12.75 |
3 |
5 |
0 |
87 |
391.448 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.10 |
4.63 |
-53.64 |
2 |
5 |
-1 |
89 |
390.44 |
4 |
↓
|
|
|
|
Analogs
-
33838895
-
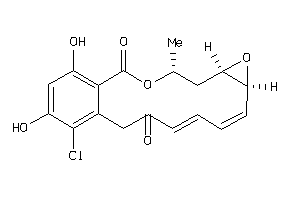
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBR1-1-E |
Carbonyl Reductase [NADPH] 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.35 |
Binding ≤ 10μM
|
|
HS90A-1-E |
Heat Shock Protein HSP 90-alpha (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
530 |
0.35 |
Binding ≤ 10μM |
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.43 |
Binding ≤ 10μM
|
|
PGH2-7-E |
Cyclooxygenase-2 (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
27 |
0.42 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
180 |
0.38 |
Binding ≤ 10μM
|
|
HSC82-1-F |
Heat Shock Protein HSP 90 (HSC82) (cluster #1 Of 1), Fungal |
Fungi |
200 |
0.38 |
Binding ≤ 10μM
|
|
HSP82-1-F |
Heat Shock Protein HSP 90 (HSP82) (cluster #1 Of 1), Fungal |
Fungi |
19 |
0.43 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7200 |
0.29 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
200 |
0.38 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
89 |
0.39 |
Functional ≤ 10μM
|
|
Z80339-1-O |
NRK (cluster #1 Of 1), Other |
Other |
290 |
0.37 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
38 |
0.42 |
Functional ≤ 10μM
|
|
Z80622-1-O |
3Y1 Cell Line (cluster #1 Of 1), Other |
Other |
700 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
3.67 |
-13.53 |
2 |
6 |
0 |
96 |
364.781 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.70 |
5.33 |
-120.39 |
0 |
6 |
-2 |
102 |
362.765 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.70 |
4.33 |
-46.69 |
1 |
6 |
-1 |
99 |
363.773 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
2.15 |
-9.78 |
2 |
5 |
0 |
79 |
342.298 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
7.19 |
-13.47 |
2 |
7 |
0 |
92 |
318.768 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.12 |
7.45 |
-39.43 |
3 |
7 |
1 |
93 |
319.776 |
3 |
↓
|
|
|
|
Analogs
-
4655391
-
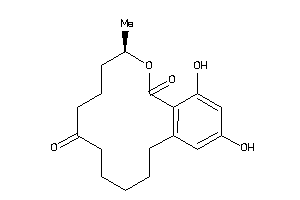
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.43 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
5.85 |
-9.17 |
2 |
5 |
0 |
84 |
320.385 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.55 |
6.86 |
-43.81 |
1 |
5 |
-1 |
87 |
319.377 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1560 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.11 |
9.15 |
-6.77 |
2 |
5 |
0 |
77 |
344.205 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
5.11 |
7.77 |
-6.42 |
2 |
5 |
0 |
77 |
344.205 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
1.93 |
-11.14 |
6 |
7 |
0 |
141 |
353.407 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.35 |
Binding ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
23 |
0.32 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
3 |
0.36 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
53 |
0.31 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
17 |
0.33 |
Functional ≤ 10μM
|
|
Z80488-1-O |
SK-MEL-5 (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
18 |
0.33 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
3 |
0.36 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
3 |
0.36 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
52 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.72 |
5.26 |
-20.38 |
4 |
7 |
0 |
110 |
464.488 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.28 |
9.87 |
-38.97 |
4 |
7 |
1 |
95 |
481.429 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.28 |
7.36 |
-9.31 |
3 |
7 |
0 |
93 |
480.421 |
7 |
↓
|
|
|
|
Analogs
-
5190651
-
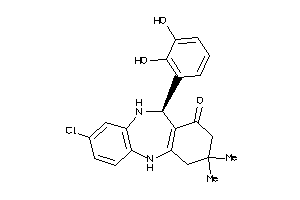
-
5190778
-
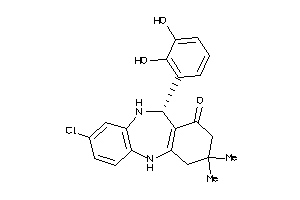
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-1-E |
Heat Shock Protein HSP 90-alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.30 |
Functional ≤ 10μM
|
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
4.9 |
-10.83 |
4 |
5 |
0 |
82 |
350.418 |
1 |
↓
|
|
|
|
Analogs
-
5190651
-

-
5190778
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-1-E |
Heat Shock Protein HSP 90-alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.30 |
Functional ≤ 10μM
|
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
4.15 |
-11.9 |
4 |
5 |
0 |
82 |
350.418 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.37 |
Functional ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
4.53 |
-17.37 |
2 |
5 |
0 |
80 |
288.23 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.90 |
5.01 |
-38.27 |
1 |
5 |
-1 |
83 |
287.222 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.90 |
5.09 |
-39.75 |
1 |
5 |
-1 |
83 |
287.222 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.39 |
Functional ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.55 |
1.17 |
-17.07 |
3 |
6 |
0 |
103 |
292.316 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.01 |
0.12 |
-94.02 |
1 |
6 |
-2 |
109 |
290.3 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.55 |
2.1 |
-46.41 |
2 |
6 |
-1 |
106 |
291.308 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-3-E |
Heat Shock Protein HSP 90-alpha (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.44 |
Functional ≤ 10μM
|
|
HS90B-3-E |
Heat Shock Protein HSP 90-beta (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
2.1 |
-8.9 |
2 |
4 |
0 |
66 |
283.736 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.09 |
2.86 |
-40.38 |
1 |
4 |
-1 |
69 |
282.728 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.41 |
Functional ≤ 10μM
|
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
3.8 |
-10.73 |
2 |
4 |
0 |
58 |
272.329 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-3-E |
Heat Shock Protein HSP 90-alpha (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.54 |
Functional ≤ 10μM
|
|
HS90B-3-E |
Heat Shock Protein HSP 90-beta (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.23 |
3.33 |
-9.02 |
1 |
4 |
0 |
49 |
210.62 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-1-E |
Heat Shock Protein HSP 90-alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Functional ≤ 10μM
|
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
6.41 |
-11.23 |
3 |
5 |
0 |
77 |
289.389 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.64 |
5.83 |
-39.89 |
4 |
5 |
1 |
78 |
290.397 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.64 |
6.81 |
-30.71 |
4 |
5 |
1 |
78 |
290.397 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-3-E |
Heat Shock Protein HSP 90-alpha (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Functional ≤ 10μM
|
|
HS90B-3-E |
Heat Shock Protein HSP 90-beta (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
7.51 |
-11.15 |
1 |
6 |
0 |
62 |
365.433 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.60 |
8.14 |
-38.85 |
2 |
6 |
1 |
63 |
366.441 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.60 |
8.15 |
-38.89 |
2 |
6 |
1 |
63 |
366.441 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-3-E |
Heat Shock Protein HSP 90-alpha (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2500 |
0.41 |
Functional ≤ 10μM
|
|
HS90B-3-E |
Heat Shock Protein HSP 90-beta (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2500 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
-1.13 |
-13.3 |
2 |
4 |
0 |
58 |
257.289 |
3 |
↓
|
|