|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-2-E |
Kynurenine 3-monooxygenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
6.41 |
-52.4 |
0 |
9 |
-1 |
125 |
420.448 |
7 |
↓
|
|
|
|
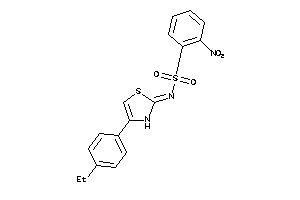
Analogs
-
5924378
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-2-E |
Kynurenine 3-monooxygenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
48 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
7.88 |
-53.2 |
0 |
7 |
-1 |
107 |
374.423 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.74 |
7.59 |
-18.43 |
1 |
7 |
0 |
108 |
375.431 |
4 |
↓
|
|
|
|
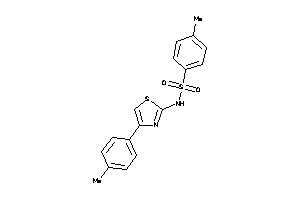
Analogs
-
475636
-
-
4951251
-
-
5392779
-
-
6707041
-
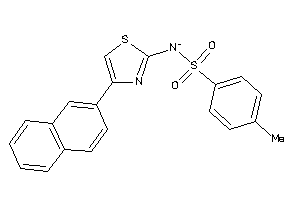
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-2-E |
Kynurenine 3-monooxygenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
470 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
7.19 |
-49.32 |
0 |
4 |
-1 |
61 |
329.426 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.81 |
7.42 |
-14.05 |
1 |
4 |
0 |
62 |
330.434 |
3 |
↓
|
|
|
|
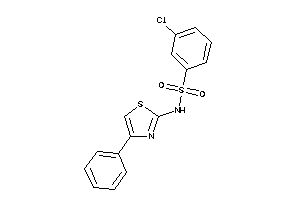
Analogs
-
2580311
-
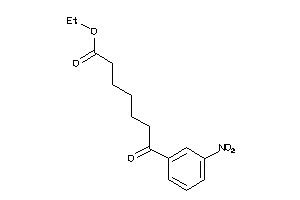
-
2580312
-
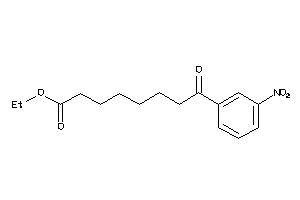
-
38812520
-
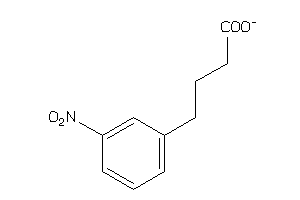
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5770 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.18 |
6.2 |
-49.36 |
0 |
6 |
-1 |
103 |
222.176 |
5 |
↓
|
|
|
|
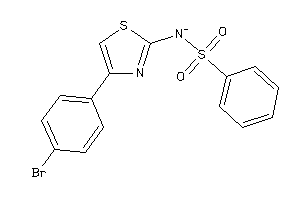
Analogs
-
5392779
-

-
6707041
-

-
193791
-
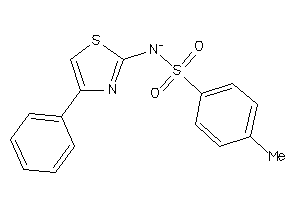
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-2-E |
Kynurenine 3-monooxygenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.25 |
8.1 |
-13.95 |
1 |
4 |
0 |
62 |
344.461 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.33 |
7.86 |
-49.38 |
0 |
4 |
-1 |
61 |
343.453 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
950 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
8.5 |
-44.75 |
0 |
4 |
-1 |
74 |
260.052 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
7.17 |
-42.16 |
0 |
4 |
-1 |
66 |
274.079 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.05 |
3.19 |
-13.63 |
0 |
4 |
0 |
60 |
275.087 |
5 |
↓
|
|
|
|
Analogs
-
6092539
-
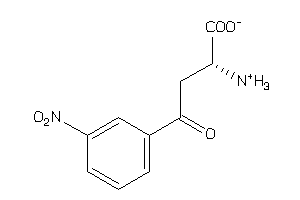
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.52 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.68 |
-1.2 |
-41.39 |
3 |
7 |
0 |
130 |
238.199 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
8.04 |
-46.28 |
0 |
4 |
-1 |
74 |
225.607 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.76 |
6.32 |
-42.26 |
0 |
4 |
-1 |
66 |
241.169 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.02 |
6.31 |
-13.82 |
0 |
4 |
0 |
60 |
242.177 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.16 |
6.71 |
-46.02 |
0 |
4 |
-1 |
66 |
239.634 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.42 |
7.98 |
-13.77 |
0 |
4 |
0 |
60 |
240.642 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
8.62 |
-46.36 |
0 |
4 |
-1 |
74 |
239.634 |
4 |
↓
|
|
|
|
Analogs
-
2566916
-
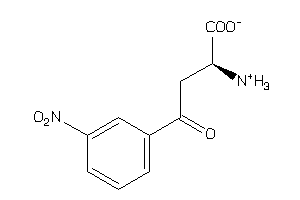
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.68 |
-1.2 |
-42.02 |
3 |
7 |
0 |
130 |
238.199 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
7.94 |
-37.7 |
0 |
4 |
-1 |
66 |
257.624 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.53 |
6.76 |
-13.16 |
0 |
4 |
0 |
60 |
258.632 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
6.82 |
-43.29 |
0 |
4 |
-1 |
66 |
284.085 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.76 |
6.96 |
-42.01 |
0 |
4 |
-1 |
66 |
284.085 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.57 |
1.06 |
-14.29 |
0 |
4 |
0 |
60 |
285.093 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
7.45 |
-42.21 |
0 |
4 |
-1 |
66 |
223.179 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.90 |
6.27 |
-17.3 |
0 |
4 |
0 |
60 |
224.187 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
6.81 |
-45.9 |
0 |
4 |
-1 |
66 |
284.085 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.55 |
6.81 |
-16.35 |
0 |
4 |
0 |
60 |
285.093 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
6145349
-
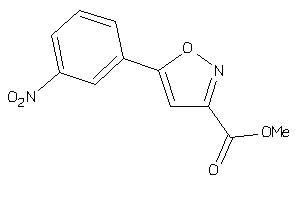
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-2-E |
Kynurenine 3-monooxygenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2680 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
5.87 |
-52.87 |
0 |
7 |
-1 |
112 |
233.159 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-2-E |
Kynurenine 3-monooxygenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
240 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.86 |
6.72 |
-15.49 |
1 |
5 |
0 |
72 |
360.46 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.94 |
6.49 |
-48.54 |
0 |
5 |
-1 |
70 |
359.452 |
5 |
↓
|
|
|
|
Analogs
-
33933542
-
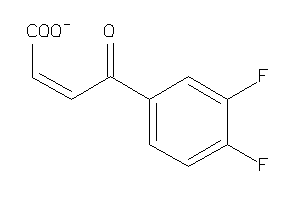
-
36180248
-
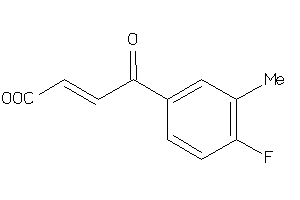
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.48 |
5.38 |
-44.76 |
0 |
3 |
-1 |
57 |
211.143 |
3 |
↓
|
|
|
|
Analogs
-
33884549
-
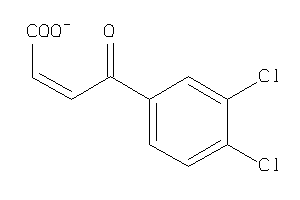
-
36179835
-
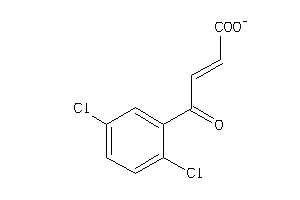
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
0.99 |
-44.08 |
0 |
3 |
-1 |
57 |
244.053 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
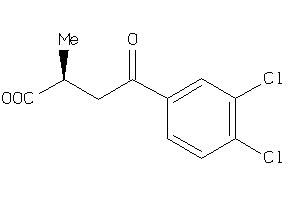
Analogs
-
22202
-
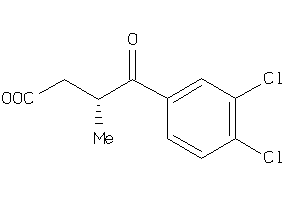
-
22203
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KMO-1-E |
Kynurenine 3-monooxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
7.11 |
-44.09 |
0 |
3 |
-1 |
57 |
260.096 |
4 |
↓
|
|