|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
62 |
0.53 |
Binding ≤ 10μM
|
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
215 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
9.45 |
-7.77 |
0 |
5 |
0 |
55 |
266.341 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
314 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.91 |
8.28 |
-45.26 |
1 |
6 |
1 |
60 |
268.337 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.91 |
5.89 |
-7.35 |
0 |
6 |
0 |
59 |
267.329 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
180 |
0.47 |
Binding ≤ 10μM
|
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
5.55 |
-11.04 |
1 |
6 |
0 |
76 |
282.34 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
180 |
0.47 |
Binding ≤ 10μM
|
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
5.59 |
-11.1 |
1 |
6 |
0 |
76 |
282.34 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
49 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
9.88 |
-11.87 |
0 |
7 |
0 |
82 |
324.377 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CTRA-2-E |
Alpha-chymotrypsin (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.75 |
Binding ≤ 10μM
|
|
LIPS-2-E |
Hormone Sensitive Lipase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.52 |
Binding ≤ 10μM
|
|
MDHC-2-E |
Malate Dehydrogenase Cytoplasmic (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.67 |
Binding ≤ 10μM
|
|
MDHM-2-E |
Malate Dehydrogenase, Mitochondrial (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.71 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
9.11 |
-6.44 |
0 |
2 |
0 |
26 |
212.248 |
4 |
↓
|
|
|
|
Analogs
-
34395899
-
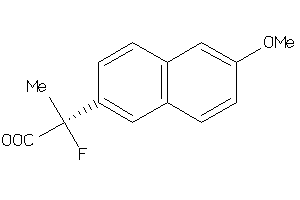
Draw
Identity
99%
90%
80%
70%
Vendors
And 89 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.38 |
7.87 |
-47.82 |
0 |
3 |
-1 |
49 |
229.255 |
3 |
↓
|
|
|
|
Analogs
-
5849525
-
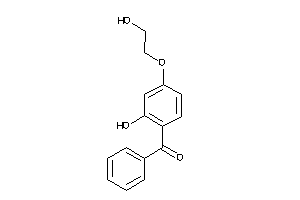
-
187
-
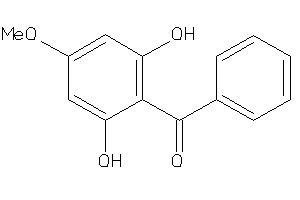
Draw
Identity
99%
90%
80%
70%
Vendors
And 40 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-2-E |
Hormone Sensitive Lipase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3250 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
4.46 |
-8.1 |
1 |
3 |
0 |
47 |
228.247 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.37 |
5.24 |
-49.88 |
0 |
3 |
-1 |
49 |
227.239 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
166 |
0.50 |
Binding ≤ 10μM
|
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
450 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.05 |
4.96 |
-10.43 |
1 |
6 |
0 |
76 |
268.313 |
1 |
↓
|
|
|
|
Analogs
-
2388308
-
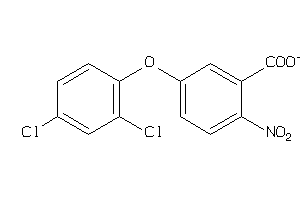
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-3-E |
Hormone Sensitive Lipase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
430 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.21 |
10.08 |
-10.95 |
0 |
6 |
0 |
81 |
342.134 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-1-E |
Hormone Sensitive Lipase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
2.84 |
-12.58 |
1 |
8 |
0 |
99 |
328.328 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-3-E |
Hormone Sensitive Lipase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
370 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
5.26 |
-9.19 |
1 |
3 |
0 |
38 |
225.313 |
5 |
↓
|
|