|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.45 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.37 |
9.96 |
-14.78 |
1 |
4 |
0 |
43 |
348.446 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
-0.43 |
-12.34 |
1 |
3 |
0 |
38 |
243.306 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
243 |
0.54 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
243 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
3.55 |
-11.93 |
2 |
3 |
0 |
49 |
229.279 |
3 |
↓
|
|
|
|
Analogs
-
7031
-
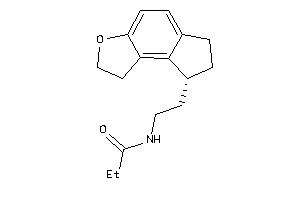
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
6.64 |
-10.06 |
1 |
3 |
0 |
38 |
259.349 |
4 |
↓
|
|
|
|
Analogs
-
4783806
-
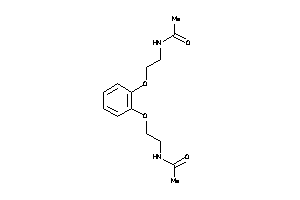
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1B-2-E |
Melatonin Receptor 1B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6607 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
3.02 |
-11.09 |
1 |
4 |
0 |
48 |
209.245 |
5 |
↓
|
|
|
|
Analogs
-
42896551
-
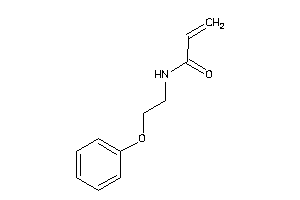
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1B-2-E |
Melatonin Receptor 1B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5495 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
3.13 |
-8.79 |
1 |
3 |
0 |
38 |
179.219 |
4 |
↓
|
|
|
|
Analogs
-
37855011
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-2-E |
Melatonin Receptor 1A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3802 |
0.58 |
Binding ≤ 10μM
|
|
MTR1B-2-E |
Melatonin Receptor 1B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
339 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
4.73 |
-10.04 |
1 |
2 |
0 |
29 |
195.287 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
-1.53 |
-14 |
2 |
4 |
0 |
54 |
246.31 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
5.7 |
-13.41 |
2 |
4 |
0 |
54 |
272.348 |
5 |
↓
|
|
|
|
Analogs
-
34589727
-
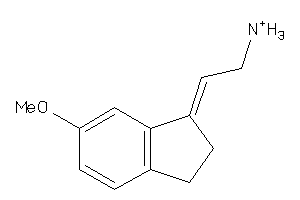
-
34596901
-
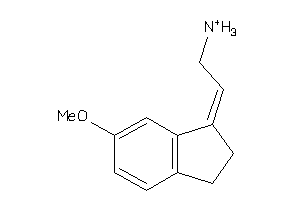
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
NQO2-3-E |
Quinone Reductase 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1190 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
6.09 |
-9.42 |
1 |
3 |
0 |
38 |
245.322 |
4 |
↓
|
|
|
|
Analogs
-
3960338
-
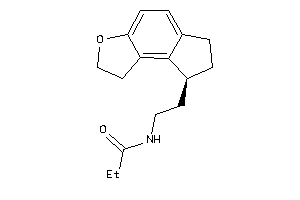
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
6.52 |
-10.63 |
1 |
3 |
0 |
38 |
259.349 |
4 |
↓
|
|
|
|
Analogs
-
34589727
-

-
34596901
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.70 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.70 |
Binding ≤ 10μM
|
|
NQO2-3-E |
Quinone Reductase 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
467 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
5.86 |
-12.4 |
1 |
3 |
0 |
38 |
245.322 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1C-1-E |
Melatonin Receptor 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
6.99 |
-13.76 |
2 |
4 |
0 |
54 |
308.381 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
10.25 |
-12.84 |
0 |
4 |
0 |
39 |
339.435 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
9.7 |
-11.15 |
0 |
4 |
0 |
39 |
339.435 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.63 |
Binding ≤ 10μM
|
|
Z81346-1-O |
Melanophores (Melanophore Cells) (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
4.64 |
-11.02 |
2 |
4 |
0 |
54 |
260.337 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-2-E |
Melatonin Receptor 1A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
84 |
0.52 |
Binding ≤ 10μM
|
|
MTR1B-2-E |
Melatonin Receptor 1B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
4.99 |
-12.58 |
2 |
4 |
0 |
50 |
256.305 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
66 |
0.36 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
11.15 |
-14.07 |
1 |
4 |
0 |
43 |
376.5 |
6 |
↓
|
|
|
|
Analogs
-
34544038
-
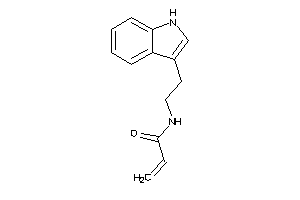
-
36612220
-
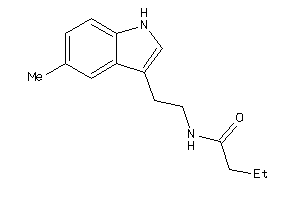
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
719 |
0.54 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
719 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
5 |
-12.36 |
2 |
3 |
0 |
45 |
216.284 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
73 |
0.62 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
73 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.58 |
4.4 |
-13.48 |
2 |
3 |
0 |
45 |
220.247 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
3995799
-
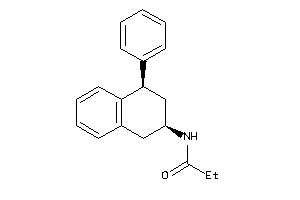
-
14092
-
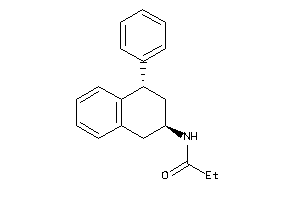
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
59 |
0.48 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
9.46 |
-8.55 |
1 |
2 |
0 |
29 |
279.383 |
3 |
↓
|
|
|
|
Analogs
-
14092
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
59 |
0.48 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
9.45 |
-8.56 |
1 |
2 |
0 |
29 |
279.383 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1606 |
0.37 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1606 |
0.37 |
Binding ≤ 10μM
|
|
MTR1C-1-E |
Melatonin Receptor 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1606 |
0.37 |
Binding ≤ 10μM
|
|
Z81346-1-O |
Melanophores (Melanophore Cells) (cluster #1 Of 1), Other |
Other |
2100 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
8.44 |
-12.45 |
2 |
3 |
0 |
45 |
292.382 |
5 |
↓
|
|
|
|
Analogs
-
36612210
-
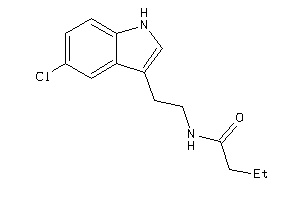
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.66 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
-1.76 |
-12.62 |
2 |
3 |
0 |
45 |
236.702 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
4.5 |
-11.69 |
2 |
4 |
0 |
54 |
358.179 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.41 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
7.6 |
-12.34 |
1 |
3 |
0 |
34 |
242.322 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-2-E |
Melatonin Receptor 1A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.72 |
Binding ≤ 10μM
|
|
MTR1B-2-E |
Melatonin Receptor 1B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.72 |
Binding ≤ 10μM
|
|
MTR1C-1-E |
Melatonin Receptor 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.72 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
5.17 |
-9.63 |
1 |
3 |
0 |
38 |
221.3 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.55 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.33 |
-0.61 |
-19.46 |
2 |
7 |
0 |
99 |
277.28 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
6.17 |
-12.25 |
1 |
4 |
0 |
43 |
372.206 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1060 |
0.42 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.43 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.33 |
4.32 |
-22.55 |
2 |
7 |
0 |
100 |
277.28 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MT3-1-E |
Metallothionein-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1880 |
0.36 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.47 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.61 |
6.91 |
-19.59 |
1 |
7 |
0 |
89 |
417.203 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
617 |
0.41 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
617 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.22 |
3.51 |
-17.33 |
2 |
6 |
0 |
73 |
292.335 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.89 |
-14.01 |
1 |
3 |
0 |
38 |
305.805 |
7 |
↓
|
|
|
|
Analogs
-
34021978
-
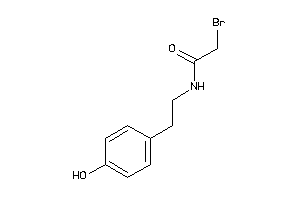
-
41725161
-
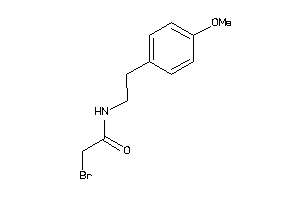
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
6.43 |
-12.86 |
1 |
3 |
0 |
38 |
322.202 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
0.23 |
-11.21 |
2 |
4 |
0 |
54 |
286.253 |
5 |
↓
|
|
|
|
Analogs
-
10146635
-
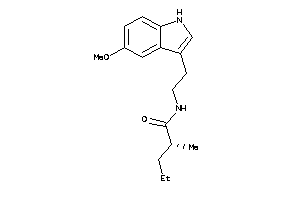
-
10146636
-
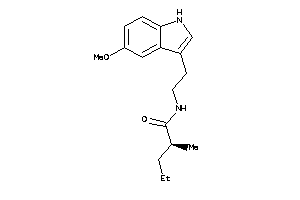
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
-1.82 |
-13.89 |
2 |
4 |
0 |
54 |
272.348 |
5 |
↓
|
|
|
|
Analogs
-
2137649
-
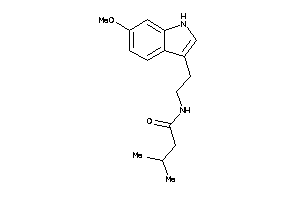
-
2137842
-
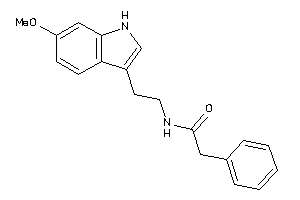
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4800 |
0.44 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4800 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
3.63 |
-13.49 |
2 |
4 |
0 |
54 |
232.283 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
-2.59 |
-11.94 |
2 |
4 |
0 |
54 |
311.179 |
4 |
↓
|
|
|
|
Analogs
-
3825731
-
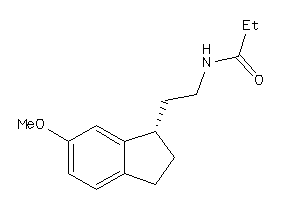
-
34629121
-
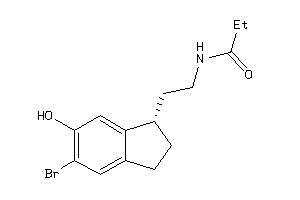
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
-0.59 |
-9.43 |
1 |
3 |
0 |
38 |
247.338 |
5 |
↓
|
|
|
|
Analogs
-
34629121
-

-
21169
-
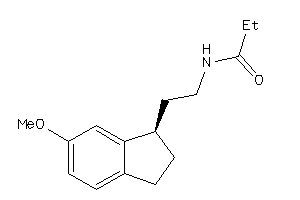
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
NQO2-4-E |
Quinone Reductase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
686 |
0.48 |
Binding ≤ 10μM
|
|
NQO2-4-E |
Quinone Reductase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3570 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
-0.59 |
-9.41 |
1 |
3 |
0 |
38 |
247.338 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
4.17 |
-14.22 |
2 |
4 |
0 |
54 |
266.728 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.64 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
5.2 |
-13.91 |
2 |
4 |
0 |
54 |
258.321 |
5 |
↓
|
|
|
|
Analogs
-
2995263
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-2-E |
Melatonin Receptor 1A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.69 |
Binding ≤ 10μM
|
|
MTR1B-2-E |
Melatonin Receptor 1B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
68 |
0.67 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.17 |
2.45 |
-11.81 |
1 |
4 |
0 |
48 |
209.245 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1B-2-E |
Melatonin Receptor 1B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
108 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
1.28 |
-12.6 |
1 |
4 |
0 |
47 |
285.343 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 57 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MT3-1-E |
Metallothionein-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.59 |
Binding ≤ 10μM
|
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.74 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.74 |
Binding ≤ 10μM
|
|
MTR1C-1-E |
Melatonin Receptor 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.74 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
65 |
0.59 |
Binding ≤ 10μM
|
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
23 |
0.63 |
Functional ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
23 |
0.63 |
Functional ≤ 10μM
|
|
MTR1C-1-E |
Melatonin Receptor 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
23 |
0.63 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81346-1-O |
Melanophores (Melanophore Cells) (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
3.62 |
-14.46 |
2 |
4 |
0 |
54 |
232.283 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.11 |
3.72 |
-41.44 |
2 |
4 |
1 |
52 |
233.291 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.11 |
3.72 |
-41.43 |
2 |
4 |
1 |
52 |
233.291 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
82 |
0.40 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
-1.96 |
-14.14 |
3 |
4 |
0 |
64 |
334.419 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MT3-1-E |
Metallothionein-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
58 |
0.51 |
Binding ≤ 10μM
|
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.42 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
2.54 |
-17.68 |
3 |
6 |
0 |
83 |
275.308 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.51 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.51 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
5.24 |
-18.84 |
2 |
7 |
0 |
100 |
403.176 |
5 |
↓
|
|