|
|
Analogs
-
39295391
-
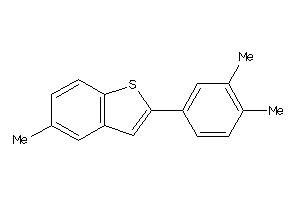
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-2-E |
Quinone Reductase 1) (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.31 |
Binding ≤ 10μM
|
|
NQO2-2-E |
Quinone Reductase 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.28 |
1.14 |
-10.3 |
0 |
0 |
0 |
0 |
360.481 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.34 |
Binding ≤ 10μM
|
|
NQO2-4-E |
Quinone Reductase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3900 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.31 |
5.86 |
-7.65 |
3 |
4 |
0 |
65 |
304.349 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.31 |
6.64 |
-54.23 |
2 |
4 |
-1 |
68 |
303.341 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.31 |
6.63 |
-48.16 |
2 |
4 |
-1 |
68 |
303.341 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
390 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.94 |
1.89 |
-8.92 |
0 |
1 |
0 |
12 |
383.491 |
2 |
↓
|
|
|
|
Analogs
-
6091777
-
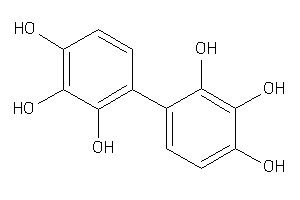
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-3-E |
Quinone Reductase 1) (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
-5.98 |
-10.11 |
5 |
5 |
0 |
101 |
234.207 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.46 |
Binding ≤ 10μM
|
|
PCSK7-1-E |
Subtilisin/kexin Type 7 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1300 |
0.33 |
Binding ≤ 10μM |
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
600 |
0.35 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
9.94 |
-141.69 |
0 |
6 |
-2 |
107 |
334.283 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
3.33 |
8.89 |
-64.51 |
1 |
6 |
-1 |
104 |
335.291 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.33 |
7.78 |
-16.67 |
2 |
6 |
0 |
101 |
336.299 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-3-E |
Quinone Reductase 1) (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
1.25 |
-4.08 |
0 |
2 |
0 |
18 |
166.22 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
3.47 |
-14.51 |
2 |
5 |
0 |
71 |
315.332 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-3-E |
Quinone Reductase 1) (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.32 |
1.67 |
-7.04 |
0 |
2 |
0 |
18 |
238.286 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-2-E |
Quinone Reductase 1) (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
10.23 |
-140.48 |
0 |
6 |
-2 |
107 |
416.41 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.42 |
10.3 |
-60.46 |
1 |
6 |
-1 |
104 |
417.418 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.42 |
8.56 |
-26.24 |
2 |
6 |
0 |
101 |
418.426 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2200 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.20 |
12.17 |
-134.57 |
0 |
6 |
-2 |
107 |
444.826 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.20 |
11.53 |
-56.53 |
1 |
6 |
-1 |
104 |
445.834 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.20 |
10.84 |
-26.71 |
2 |
6 |
0 |
101 |
446.842 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.62 |
11.68 |
-144.1 |
0 |
7 |
-2 |
110 |
453.45 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.62 |
9.76 |
-21.75 |
2 |
7 |
0 |
104 |
455.466 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.62 |
11.08 |
-63.34 |
1 |
7 |
-1 |
107 |
454.458 |
4 |
↓
|
|
|
|
Analogs
-
6143155
-
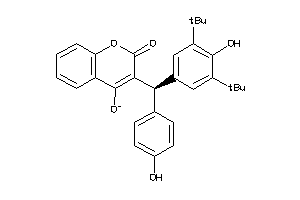
-
6143156
-
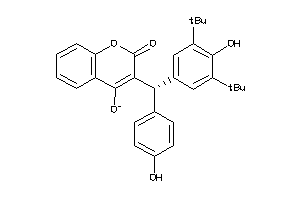
-
8453187
-
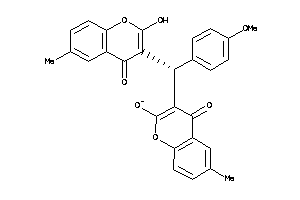
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8700 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.58 |
10.57 |
-139.4 |
0 |
7 |
-2 |
116 |
440.407 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.58 |
9.97 |
-60.78 |
1 |
7 |
-1 |
113 |
441.415 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.58 |
9.63 |
-28.94 |
2 |
7 |
0 |
110 |
442.423 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5900 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
9.83 |
-62.2 |
1 |
7 |
-1 |
117 |
401.35 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
3.78 |
10.37 |
-143.3 |
0 |
7 |
-2 |
120 |
400.342 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.78 |
9.35 |
-26.03 |
2 |
7 |
0 |
114 |
402.358 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3250 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
3.97 |
-14.46 |
2 |
3 |
0 |
56 |
161.16 |
0 |
↓
|
|
|
|
Analogs
-
6576219
-
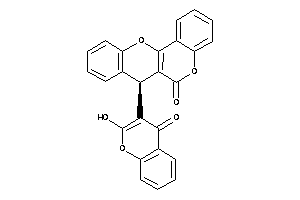
-
5835063
-
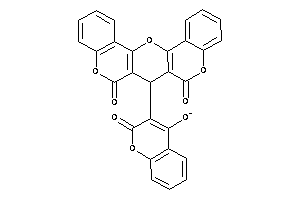
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
650 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.68 |
12.75 |
-68.03 |
0 |
6 |
-1 |
93 |
409.373 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.68 |
12.12 |
-20.01 |
1 |
6 |
0 |
90 |
410.381 |
1 |
↓
|
|
|
|
Analogs
-
5835063
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
650 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.68 |
12.76 |
-68.06 |
0 |
6 |
-1 |
93 |
409.373 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.68 |
12.13 |
-20.03 |
1 |
6 |
0 |
90 |
410.381 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
8.8 |
-139.74 |
1 |
7 |
-2 |
127 |
426.38 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.04 |
8.53 |
-66.29 |
2 |
7 |
-1 |
124 |
427.388 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.04 |
8.15 |
-60.34 |
2 |
7 |
-1 |
124 |
427.388 |
3 |
↓
|
|
|
|
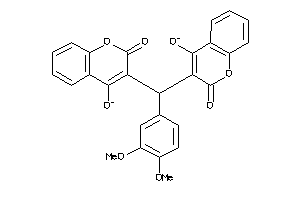
Analogs
-
11865780
-
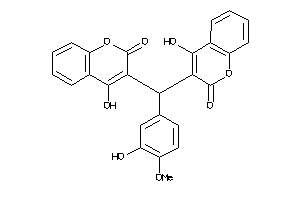
-
5225136
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
8.55 |
-140.87 |
1 |
8 |
-2 |
136 |
456.406 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.86 |
7.81 |
-61.39 |
2 |
8 |
-1 |
133 |
457.414 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.86 |
7.52 |
-60.22 |
2 |
8 |
-1 |
133 |
457.414 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
588 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.37 |
10.25 |
-59.29 |
1 |
6 |
-1 |
104 |
377.372 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.37 |
9.77 |
-140.99 |
0 |
6 |
-2 |
107 |
376.364 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.37 |
8.46 |
-35.78 |
2 |
6 |
0 |
101 |
378.38 |
4 |
↓
|
|
|
|
Analogs
-
4261867
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.08 |
-11.06 |
-49.22 |
6 |
9 |
-1 |
183 |
349.3 |
1 |
↓
|
|
|
|
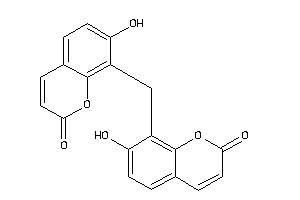
Analogs
-
641098
-

-
39289818
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5500 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
11.28 |
-143.52 |
0 |
6 |
-2 |
107 |
362.337 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.17 |
10.5 |
-64.53 |
1 |
6 |
-1 |
104 |
363.345 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.17 |
9.72 |
-19.01 |
2 |
6 |
0 |
101 |
364.353 |
2 |
↓
|
|
|
|
Analogs
-
39289818
-

-
641098
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
8.53 |
-44.95 |
0 |
3 |
-1 |
53 |
251.261 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.61 |
7.88 |
-18.45 |
1 |
3 |
0 |
50 |
252.269 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO1-1-E |
Quinone Reductase 1) (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.96 |
-6.92 |
-47.52 |
6 |
8 |
1 |
131 |
401.439 |
9 |
↓
|
|