|
|
Analogs
-
31302698
-
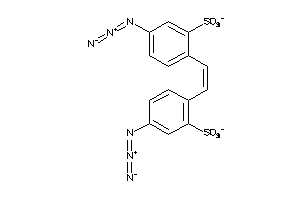
-
31302701
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
TNFB-1-E |
TNF-beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.71 |
3.05 |
-98.78 |
0 |
8 |
-2 |
139 |
452.516 |
6 |
↓
|
|
|
|
Analogs
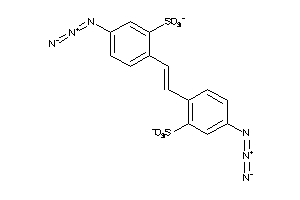
-
15637617
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.25 |
Functional ≤ 10μM
|
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.26 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
690 |
0.28 |
Functional ≤ 10μM
|
|
P2RX4-1-E |
P2X Purinoceptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.25 |
Functional ≤ 10μM
|
|
P2RX5-1-E |
P2X Purinoceptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.28 |
Functional ≤ 10μM
|
|
P2RX6-1-E |
P2X Purinoceptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.27 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.27 |
Functional ≤ 10μM
|
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1720 |
0.26 |
Functional ≤ 10μM
|
|
P2Y11-1-E |
Purinergic Receptor P2Y11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.25 |
Functional ≤ 10μM
|
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.75 |
-2.04 |
-153.5 |
6 |
17 |
-2 |
268 |
521.234 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.92 |
-1.22 |
-126.98 |
6 |
17 |
-1 |
266 |
522.242 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.92 |
-5.6 |
-128.24 |
6 |
17 |
-1 |
266 |
522.242 |
8 |
↓
|
|
|
|
Analogs
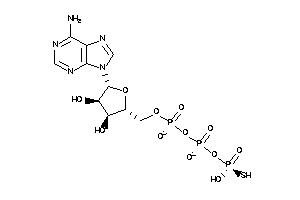
-
8295117
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
14 |
0.35 |
Binding ≤ 10μM
|
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.30 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.26 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9200 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 45 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAP2-1-E |
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.23 |
Binding ≤ 10μM
|
|
P2RY1-1-E |
Purinergic Receptor P2Y1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.25 |
Binding ≤ 10μM
|
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.33 |
Functional ≤ 10μM
|
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.26 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.29 |
Functional ≤ 10μM
|
|
P2RX4-1-E |
P2X Purinoceptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.28 |
Functional ≤ 10μM
|
|
P2RX5-1-E |
P2X Purinoceptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RX6-1-E |
P2X Purinoceptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.23 |
Functional ≤ 10μM
|
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
96 |
0.32 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.26 |
Functional ≤ 10μM
|
|
P2Y11-1-E |
Purinergic Receptor P2Y11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.25 |
Functional ≤ 10μM |
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3190 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.74 |
-1.4 |
-65.19 |
6 |
14 |
-1 |
218 |
442.263 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.92 |
0.87 |
-231.59 |
4 |
14 |
-3 |
218 |
440.247 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.74 |
-5.71 |
-75.71 |
7 |
14 |
0 |
220 |
443.271 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY1-1-E |
Purinergic Receptor P2Y1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
54 |
0.31 |
Functional ≤ 10μM
|
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.25 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.27 |
Functional ≤ 10μM
|
|
P2RX4-1-E |
P2X Purinoceptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Functional ≤ 10μM
|
|
P2RX5-1-E |
P2X Purinoceptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Functional ≤ 10μM
|
|
P2RX6-1-E |
P2X Purinoceptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.26 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.35 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Functional ≤ 10μM
|
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.06 |
-2 |
-367.11 |
4 |
18 |
-4 |
290 |
549.244 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.06 |
-3.15 |
-229.34 |
5 |
18 |
-3 |
288 |
550.252 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY1-1-E |
Purinergic Receptor P2Y1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
54 |
0.31 |
Functional ≤ 10μM
|
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.25 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.27 |
Functional ≤ 10μM
|
|
P2RX4-1-E |
P2X Purinoceptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Functional ≤ 10μM
|
|
P2RX5-1-E |
P2X Purinoceptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Functional ≤ 10μM
|
|
P2RX6-1-E |
P2X Purinoceptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.26 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.35 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Functional ≤ 10μM
|
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.31 |
-3.32 |
-290.48 |
1 |
15 |
-4 |
262 |
507.351 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.31 |
-2.32 |
-388.42 |
0 |
15 |
-5 |
265 |
506.343 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.31 |
-4.47 |
-174.22 |
2 |
15 |
-3 |
259 |
508.359 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.31 |
-3.32 |
-320.92 |
1 |
15 |
-4 |
262 |
507.351 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.31 |
-2.32 |
-422.37 |
0 |
15 |
-5 |
265 |
506.343 |
8 |
↓
|
|