|
|
Analogs
-
13704137
-
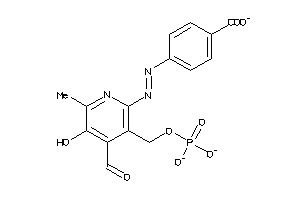
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.42 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.63 |
4.76 |
-211.02 |
1 |
11 |
-3 |
187 |
392.24 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.63 |
5.76 |
-289.42 |
0 |
11 |
-4 |
190 |
391.232 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-Amino-4-(2'-(4",6"-dichloro-s-triazin-2-yl)amino)phenylamino)9,10-dihydro-9,10-dioxoanthracene-2,4'-disulphonic acid; 2-Anthracenesulfonic acid, 1-amino-4-((3-((4,6-dichloro-1,3,5-triazin-2-yl)amino)-4-sulfophenyl)amino)-9,10-dihydro-9,10-dioxo-; 2-Anth
1-Amino-4-(2'-(4",6"-dichloro-s-…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
422 |
0.22 |
Functional ≤ 10μM |
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
79 |
0.24 |
Functional ≤ 10μM |
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
494 |
0.22 |
Functional ≤ 10μM |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.17 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.56 |
4.68 |
-94.13 |
4 |
14 |
-2 |
237 |
635.423 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.45 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
3.83 |
-9.3 |
5 |
8 |
0 |
123 |
474.299 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.88 |
4.2 |
-33.26 |
6 |
8 |
1 |
124 |
475.307 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
4.67 |
-10.73 |
4 |
6 |
0 |
96 |
274.324 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.39 |
5.17 |
-38.05 |
5 |
6 |
1 |
98 |
275.332 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1995 |
0.36 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
126 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
5.87 |
-10.87 |
4 |
6 |
0 |
96 |
302.378 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.98 |
2.46 |
-35.89 |
5 |
6 |
1 |
98 |
303.386 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
398 |
0.45 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1585 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
4.89 |
-6 |
4 |
6 |
0 |
96 |
274.324 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.35 |
5.38 |
-32.9 |
5 |
6 |
1 |
98 |
275.332 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
79 |
0.47 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
13 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
6.02 |
-6.91 |
4 |
6 |
0 |
96 |
400.22 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.36 |
6.51 |
-32.33 |
5 |
6 |
1 |
98 |
401.228 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.31 |
-3.32 |
-290.48 |
1 |
15 |
-4 |
262 |
507.351 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.31 |
-2.32 |
-388.42 |
0 |
15 |
-5 |
265 |
506.343 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.31 |
-4.47 |
-174.22 |
2 |
15 |
-3 |
259 |
508.359 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.31 |
-3.32 |
-320.92 |
1 |
15 |
-4 |
262 |
507.351 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.31 |
-2.32 |
-422.37 |
0 |
15 |
-5 |
265 |
506.343 |
8 |
↓
|
|