|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.47 |
Binding ≤ 10μM
|
|
Z80362-3-O |
P388 (Lymphoma Cells) (cluster #3 Of 8), Other |
Other |
125 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
5.53 |
-51.77 |
2 |
5 |
1 |
53 |
295.366 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.01 |
3.06 |
-13.85 |
1 |
5 |
0 |
52 |
294.358 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.88 |
4.61 |
-20.59 |
2 |
11 |
0 |
133 |
459.532 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
620 |
0.41 |
Binding ≤ 10μM |
|
TNKS1-1-E |
Tankyrase-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.54 |
Binding ≤ 10μM |
|
TNKS2-1-E |
Tankyrase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.56 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
7.59 |
-10.42 |
1 |
3 |
0 |
46 |
312.316 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.85 |
6.7 |
-39.29 |
0 |
3 |
-1 |
49 |
311.308 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.47 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.55 |
5.17 |
-52.45 |
5 |
5 |
1 |
88 |
325.367 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.55 |
5.17 |
-52.77 |
5 |
5 |
1 |
88 |
325.367 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.50 |
Binding ≤ 10μM
|
|
PARP2-2-E |
Poly [ADP-ribose] Polymerase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
|
PARP3-1-E |
Poly [ADP-ribose] Polymerase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.34 |
Binding ≤ 10μM
|
|
PARP4-1-E |
Poly [ADP-ribose] Polymerase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
330 |
0.38 |
Binding ≤ 10μM
|
|
TNKS1-1-E |
Tankyrase-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
6.15 |
-60.48 |
4 |
5 |
1 |
78 |
321.404 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
6.15 |
-60.47 |
4 |
5 |
1 |
78 |
321.404 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
9.75 |
-58.72 |
4 |
8 |
1 |
94 |
390.471 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.02 |
7.54 |
-14.72 |
3 |
8 |
0 |
93 |
389.463 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-3-E |
Poly [ADP-ribose] Polymerase 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
0.44 |
-9.59 |
2 |
3 |
0 |
56 |
163.176 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-3-E |
Poly [ADP-ribose] Polymerase 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
0.44 |
-9.62 |
2 |
3 |
0 |
56 |
163.176 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
68 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
-0.29 |
-45.57 |
3 |
4 |
1 |
57 |
295.362 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.77 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
3.27 |
-13.46 |
1 |
2 |
0 |
33 |
224.057 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.52 |
1.02 |
-7.07 |
2 |
5 |
0 |
72 |
279.336 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
99 |
0.75 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.29 |
-3.35 |
-12.64 |
3 |
4 |
0 |
71 |
175.191 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.69 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.69 |
Binding ≤ 10μM
|
|
Q8CFB8-1-E |
ADP-ribosyltransferase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
0.61 |
-11.07 |
2 |
4 |
0 |
66 |
176.175 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
0.80 |
0.77 |
-41.66 |
1 |
4 |
-1 |
69 |
175.167 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.80 |
1.37 |
-48.29 |
1 |
4 |
-1 |
69 |
175.167 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
1.14 |
-20.49 |
4 |
5 |
0 |
92 |
253.261 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
1.23 |
-11.91 |
3 |
4 |
0 |
72 |
229.161 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.20 |
0.75 |
-46.32 |
2 |
4 |
-1 |
70 |
228.153 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.20 |
0.78 |
-46.52 |
2 |
4 |
-1 |
70 |
228.153 |
2 |
↓
|
|
|
|
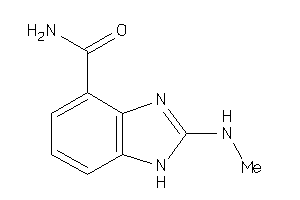
Analogs
-
36459273
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-3-E |
Poly [ADP-ribose] Polymerase 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
95 |
0.82 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.20 |
1.07 |
-18.7 |
3 |
4 |
0 |
72 |
161.164 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
1.85 |
-20.69 |
5 |
5 |
0 |
98 |
252.277 |
2 |
↓
|
|
|
|
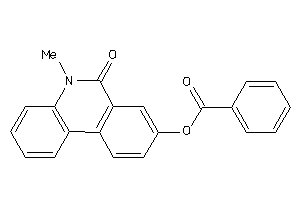
Analogs
-
39123625
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.38 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
7.92 |
-13.07 |
1 |
4 |
0 |
59 |
265.268 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
5.02 |
-121.59 |
0 |
7 |
-2 |
93 |
314.308 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.15 |
4.25 |
-13.93 |
2 |
7 |
0 |
92 |
316.324 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.15 |
5.01 |
-57.46 |
1 |
7 |
-1 |
95 |
315.316 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.33 |
Binding ≤ 10μM
|
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
4 |
-18.17 |
2 |
7 |
0 |
92 |
439.537 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
180 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.63 |
3.55 |
-12.05 |
3 |
3 |
0 |
59 |
210.236 |
0 |
↓
|
|
|
|
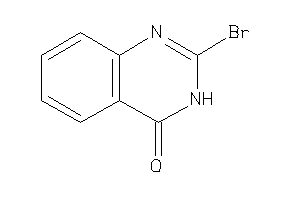
Analogs
-
34604779
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 83 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9500 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.99 |
3.46 |
-10.77 |
1 |
3 |
0 |
46 |
146.149 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.44 |
1.4 |
-44.99 |
0 |
3 |
-1 |
49 |
145.141 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 45 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-3-E |
Poly [ADP-ribose] Polymerase 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5400 |
0.82 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
1.27 |
-8.69 |
2 |
2 |
0 |
43 |
121.139 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
312 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
3.88 |
-11.48 |
3 |
3 |
0 |
59 |
210.236 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
390 |
0.75 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.23 |
0.92 |
-10.43 |
2 |
3 |
0 |
53 |
161.16 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-3-E |
Poly [ADP-ribose] Polymerase 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.66 |
Binding ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
180 |
0.59 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
1.67 |
-13.39 |
3 |
4 |
0 |
76 |
212.208 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
4.65 |
-21.08 |
3 |
7 |
0 |
118 |
282.259 |
3 |
↓
|
|
|
|
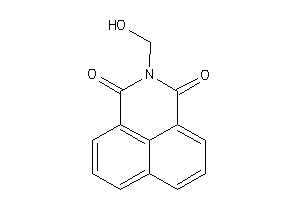
Analogs
-
34544427
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-3-E |
Poly [ADP-ribose] Polymerase 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
3.7 |
-11.07 |
1 |
3 |
0 |
50 |
197.193 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 32 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
1.02 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
0.91 |
-7.28 |
1 |
2 |
0 |
33 |
145.161 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
3,4-Dihydropyrrolo(3,2,1-jk)(1,4)benzodiazepin-1(2H)-one; BRN 0643845; LS-139234; Pyrrolo(3,2,1-jk)(1,4)benzodiazepin-1(2H)-one, 3,4-dihydro-
3,4-Dihydropyrrolo(3,2,1-jk)(1,4…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
105 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.23 |
-1.67 |
-11.93 |
1 |
3 |
0 |
34 |
186.214 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.49 |
Binding ≤ 10μM
|
|
PARP3-1-E |
Poly [ADP-ribose] Polymerase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.13 |
-0.77 |
-51.13 |
3 |
5 |
1 |
66 |
296.35 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
PARP3-1-E |
Poly [ADP-ribose] Polymerase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
-1.95 |
-50.61 |
3 |
6 |
1 |
82 |
381.431 |
3 |
↓
|
|
|
|
Analogs
-
5729165
-
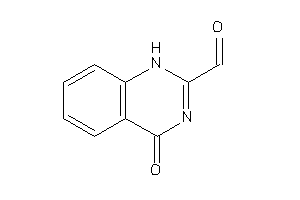
Draw
Identity
99%
90%
80%
70%
Vendors
And 71 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5600 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
3.4 |
-9.84 |
1 |
3 |
0 |
46 |
160.176 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-3-E |
Poly [ADP-ribose] Polymerase 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8000 |
0.71 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.25 |
-1.58 |
-11.05 |
3 |
3 |
0 |
63 |
137.138 |
1 |
↓
|
|
|
|
Analogs
-
32143455
-
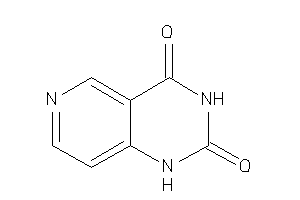
Draw
Identity
99%
90%
80%
70%
Vendors
And 74 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8100 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.63 |
0.88 |
-9.23 |
2 |
4 |
0 |
66 |
162.148 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.09 |
-1.03 |
-39.73 |
1 |
4 |
-1 |
69 |
161.14 |
0 |
↓
|
|
|
|
Analogs
-
34580013
-
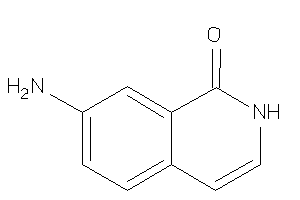
-
34580014
-
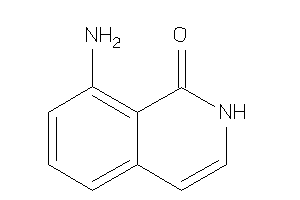
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.77 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1050 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.93 |
1.69 |
-11.1 |
3 |
3 |
0 |
59 |
160.176 |
0 |
↓
|
|
|
|
Analogs
-
3480858
-
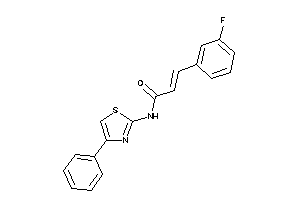
-
6344793
-
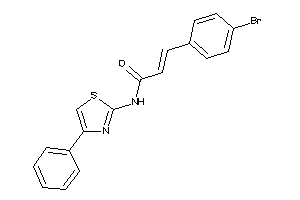
-
16771394
-
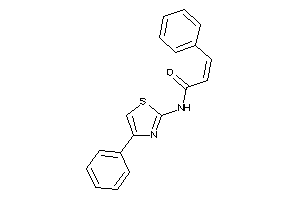
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
224 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
-0.06 |
-10.08 |
1 |
3 |
0 |
41 |
306.39 |
4 |
↓
|
|
|
|
Analogs
-
39244302
-
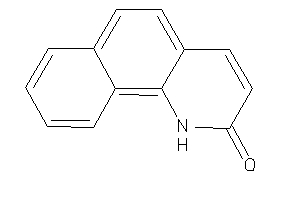
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
5.73 |
-9.7 |
1 |
2 |
0 |
33 |
195.221 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.54 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.59 |
5.27 |
-57.19 |
4 |
5 |
1 |
76 |
287.387 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.76 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
5.09 |
-11.09 |
1 |
4 |
0 |
51 |
217.272 |
0 |
↓
|
|
|
|
Analogs
-
5816056
-
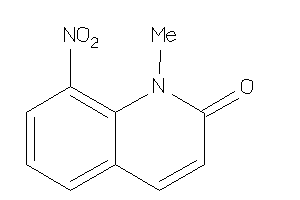
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6100 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
6.61 |
-15.57 |
1 |
5 |
0 |
79 |
240.218 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.39 |
5.79 |
-54.87 |
0 |
5 |
-1 |
82 |
239.21 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3260 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.68 |
1.39 |
-13.69 |
5 |
4 |
0 |
85 |
225.251 |
0 |
↓
|
|
|
|
Analogs
-
32222915
-
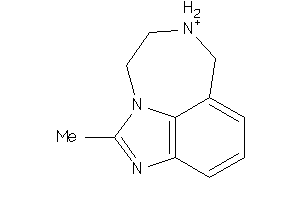
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
299 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.50 |
3.48 |
-14.13 |
1 |
4 |
0 |
47 |
187.202 |
0 |
↓
|
|
|
|
Analogs
-
36611054
-
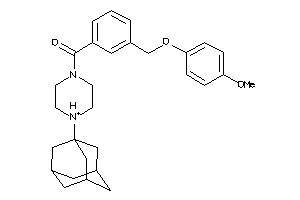
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-3-E |
Poly [ADP-ribose] Polymerase 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
188 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
9.11 |
-21.55 |
2 |
7 |
0 |
93 |
435.524 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.87 |
0.96 |
-39.95 |
5 |
5 |
1 |
88 |
233.295 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
3.17 |
-37.17 |
4 |
5 |
1 |
76 |
247.322 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
19 |
0.68 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.50 |
-0.64 |
-46.16 |
6 |
5 |
1 |
99 |
219.268 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
0.50 |
-0.67 |
-58.25 |
6 |
5 |
1 |
99 |
219.268 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.50 |
-0.92 |
-17.15 |
5 |
5 |
0 |
98 |
218.26 |
2 |
↓
|
|
|
|
Analogs
-
26507188
-
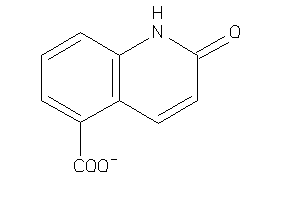
-
26507216
-
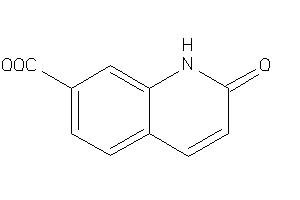
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
57 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
4.4 |
-52.88 |
1 |
4 |
-1 |
73 |
238.222 |
1 |
↓
|
|