|
|
Analogs
-
5093465
-
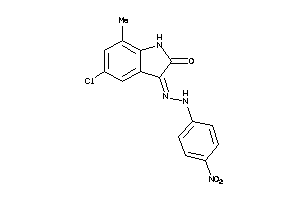
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-3-E |
Cruzipain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.45 |
Binding ≤ 10μM
|
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9400 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
3.3 |
-12.39 |
4 |
5 |
0 |
83 |
268.729 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.60 |
1.48 |
-40.21 |
3 |
5 |
-1 |
86 |
267.721 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.23 |
1.66 |
-32.4 |
4 |
5 |
0 |
86 |
268.729 |
2 |
↓
|
|
|
|
Analogs
-
39643971
-
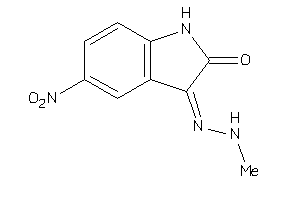
-
5342034
-
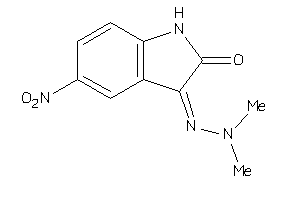
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
2.84 |
-16.85 |
4 |
8 |
0 |
129 |
265.254 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
1.05 |
2.72 |
-17.76 |
4 |
8 |
0 |
129 |
265.254 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.96 |
1.97 |
-17.27 |
4 |
8 |
0 |
137 |
265.254 |
3 |
↓
|
|
|
|
Analogs
-
4475165
-
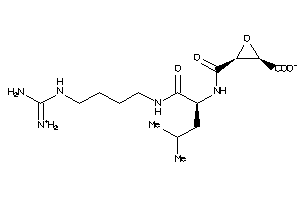
-
13493525
-
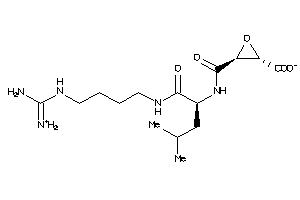
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
16 |
0.44 |
Binding ≤ 10μM
|
|
CATC-1-E |
Dipeptidyl Peptidase I (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
770 |
0.34 |
Binding ≤ 10μM
|
|
CATH-1-E |
Cathepsin H (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
PAPA1-2-E |
Papain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.41 |
Binding ≤ 10μM
|
|
Q9NBA7-1-E |
Cysteine Protease Falcipain-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
75 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
9300 |
0.28 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
2500 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.37 |
1.58 |
-72.44 |
7 |
10 |
0 |
174 |
357.411 |
12 |
↓
|
|
|
|
Analogs
-
13493525
-

-
4095645
-
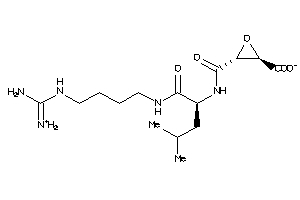
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
16 |
0.44 |
Binding ≤ 10μM
|
|
CATC-1-E |
Dipeptidyl Peptidase I (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
770 |
0.34 |
Binding ≤ 10μM
|
|
CATH-1-E |
Cathepsin H (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
PAPA1-2-E |
Papain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.41 |
Binding ≤ 10μM
|
|
Q9NBA7-1-E |
Cysteine Protease Falcipain-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
75 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
9300 |
0.28 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
2500 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.37 |
1.94 |
-73.1 |
7 |
10 |
0 |
174 |
357.411 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
1.56 |
-15.14 |
0 |
8 |
0 |
93 |
464.612 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
8.06 |
-22.37 |
3 |
9 |
0 |
122 |
469.501 |
7 |
↓
|
|
Ref
Reference (pH 7)
|
2.42 |
8.18 |
-20.55 |
3 |
9 |
0 |
122 |
469.501 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.33 |
6.66 |
-66.42 |
2 |
9 |
-1 |
128 |
468.493 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4600 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
6.34 |
-17.22 |
2 |
6 |
0 |
88 |
367.496 |
10 |
↓
|
|
|
|
Analogs
-
6635049
-
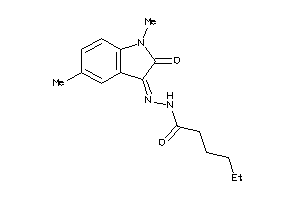
-
6635117
-
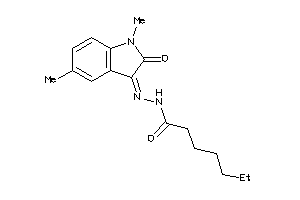
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6500 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
8.12 |
-23.05 |
2 |
10 |
0 |
127 |
460.494 |
7 |
↓
|
|
Ref
Reference (pH 7)
|
2.62 |
8.02 |
-28.67 |
2 |
10 |
0 |
127 |
460.494 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.44 |
4.25 |
-70.23 |
1 |
10 |
-1 |
137 |
459.486 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
-7.93 |
-20.2 |
1 |
6 |
0 |
97 |
482.07 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-2-E |
Falcipain 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7500 |
0.29 |
Binding ≤ 10μM
|
|
Q9NBA7-2-E |
Cysteine Protease Falcipain-3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
5.4 |
-13.97 |
3 |
6 |
0 |
91 |
359.813 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.75 |
6.17 |
-52.67 |
2 |
6 |
-1 |
94 |
358.805 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
5.4 |
-15.49 |
2 |
8 |
0 |
106 |
396.428 |
5 |
↓
|
|
|
|
Analogs
-
8084734
-
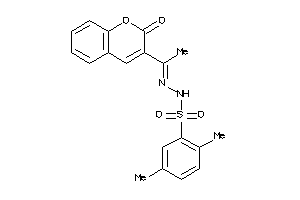
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-2-E |
Falcipain 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3900 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
5.4 |
-48.62 |
1 |
7 |
-1 |
112 |
371.394 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.05 |
5.49 |
-50.49 |
1 |
7 |
-1 |
112 |
371.394 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.05 |
5.55 |
-51.06 |
1 |
7 |
-1 |
112 |
371.394 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
4.68 |
-16.46 |
4 |
6 |
0 |
106 |
398.418 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.59 |
5.37 |
-97.83 |
2 |
6 |
-2 |
111 |
396.402 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.59 |
4.42 |
-52.78 |
3 |
6 |
-1 |
108 |
397.41 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-2-E |
Falcipain 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.29 |
Binding ≤ 10μM
|
|
Q9NBA7-2-E |
Cysteine Protease Falcipain-3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7500 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
-5.75 |
-14.84 |
4 |
6 |
0 |
105 |
382.803 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-2-E |
Falcipain 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.24 |
6.43 |
-25.68 |
4 |
8 |
0 |
123 |
402.41 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.24 |
7.43 |
-69.88 |
3 |
8 |
-1 |
126 |
401.402 |
6 |
↓
|
|
|
|
Analogs
-
4015945
-
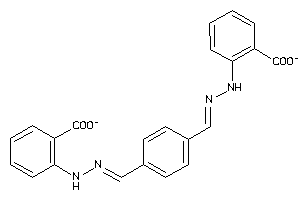
-
13458783
-
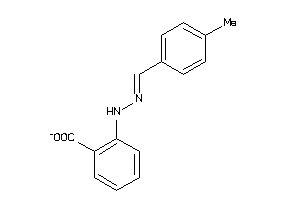
-
33246262
-
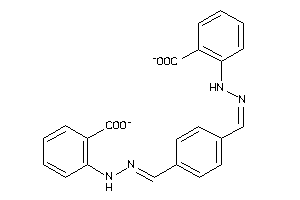
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-2-E |
Falcipain 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
|
Q9NBA7-2-E |
Cysteine Protease Falcipain-3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4900 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.68 |
12.4 |
-101.83 |
2 |
8 |
-2 |
129 |
400.394 |
8 |
↓
|
|
|
|
Analogs
-
632045
-
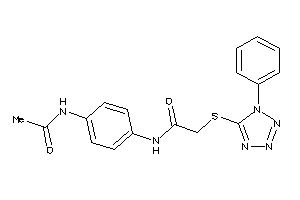
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7500 |
0.27 |
Binding ≤ 10μM
|
|
Q9NBA7-1-E |
Cysteine Protease Falcipain-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.97 |
2.73 |
-18.92 |
1 |
10 |
0 |
116 |
379.409 |
6 |
↓
|
|
|
|
Analogs
-
4401071
-
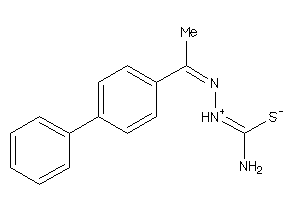
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
8.41 |
-12.09 |
3 |
3 |
0 |
50 |
269.373 |
4 |
↓
|
|
|
|
Analogs
-
1170645
-
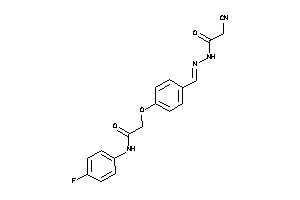
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-2-E |
Falcipain 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5700 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.87 |
7.2 |
-34.27 |
2 |
7 |
0 |
104 |
354.341 |
7 |
↓
|
|
|
|
Analogs
-
1124215
-
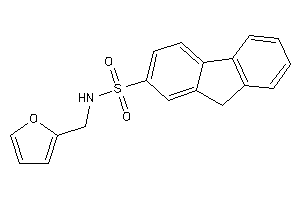
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
4.92 |
-20.13 |
2 |
8 |
0 |
119 |
484.555 |
8 |
↓
|
|
|
|
Analogs
-
1996112
-
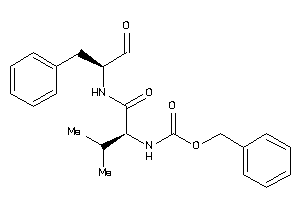
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAN1-1-E |
Calpain 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Binding ≤ 10μM
|
|
CAN2-1-E |
Calpain 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CANX-1-E |
Calpain 1/2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
25 |
0.38 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1270 |
0.29 |
Binding ≤ 10μM
|
|
CPNS1-1-E |
Calpain Small Subunit 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.37 |
Binding ≤ 10μM |
|
Q3HTL5-1-E |
Falcipain 2B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
CAN1-1-E |
Calpain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.31 |
Functional ≤ 10μM
|
|
CAN2-1-E |
Calpain 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.31 |
Functional ≤ 10μM
|
|
CPNS1-1-E |
Calpain Small Subunit 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.31 |
Functional ≤ 10μM
|
|
Z100494-1-O |
C2C12 (Myoblast Cells) (cluster #1 Of 1), Other |
Other |
10000 |
0.25 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.17 |
8.75 |
-13.99 |
2 |
6 |
0 |
85 |
382.46 |
10 |
↓
|
|
|
|
Analogs
-
1534489
-
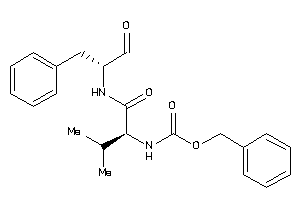
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAN1-1-E |
Calpain 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Binding ≤ 10μM
|
|
CAN2-1-E |
Calpain 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CANX-1-E |
Calpain 1/2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
25 |
0.38 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1270 |
0.29 |
Binding ≤ 10μM
|
|
CPNS1-1-E |
Calpain Small Subunit 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.37 |
Binding ≤ 10μM |
|
Q3HTL5-1-E |
Falcipain 2B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
CAN1-1-E |
Calpain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.31 |
Functional ≤ 10μM
|
|
CAN2-1-E |
Calpain 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.31 |
Functional ≤ 10μM
|
|
CPNS1-1-E |
Calpain Small Subunit 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.31 |
Functional ≤ 10μM
|
|
Z100494-1-O |
C2C12 (Myoblast Cells) (cluster #1 Of 1), Other |
Other |
10000 |
0.25 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.17 |
8.75 |
-13.21 |
2 |
6 |
0 |
85 |
382.46 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
1.16 |
-14.96 |
1 |
8 |
0 |
107 |
389.433 |
9 |
↓
|
|
|
|
|