|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 53 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
832 |
0.34 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Binding ≤ 10μM
|
|
PPARD-2-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3620 |
0.30 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.44 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.40 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
325 |
0.36 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.40 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
210 |
0.37 |
Functional ≤ 10μM
|
|
Z80106-1-O |
COS-1 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.43 |
Functional ≤ 10μM
|
|
Z80169-1-O |
Huh-7 (Hepatocellular Carcinoma) (cluster #1 Of 1), Other |
Other |
220 |
0.37 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z81117-1-O |
Keratinocytes (Keratinocytes) (cluster #1 Of 2), Other |
Other |
8000 |
0.29 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
5000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
7.4 |
-33.54 |
2 |
6 |
1 |
73 |
358.443 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.35 |
7.28 |
-11.07 |
1 |
6 |
0 |
72 |
357.435 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 54 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
832 |
0.34 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Binding ≤ 10μM
|
|
PPARD-2-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3620 |
0.30 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.44 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.41 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
325 |
0.36 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.40 |
Functional ≤ 10μM
|
|
Z80106-1-O |
COS-1 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.43 |
Functional ≤ 10μM
|
|
Z80169-1-O |
Huh-7 (Hepatocellular Carcinoma) (cluster #1 Of 1), Other |
Other |
220 |
0.37 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z81117-1-O |
Keratinocytes (Keratinocytes) (cluster #1 Of 2), Other |
Other |
8000 |
0.29 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
5000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
7.4 |
-33.23 |
2 |
6 |
1 |
73 |
358.443 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.35 |
7.28 |
-11.54 |
1 |
6 |
0 |
72 |
357.435 |
7 |
↓
|
|
|
|
Analogs
-
34648278
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
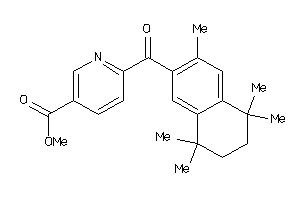
6-[1-(3,5,5,8,8-Pentamethyl-5,6,7,8-tetrahydro-naphthalen-2-yl)-cyclopropyl]-nicotinic acid
6-[1-(3,5,5,8,8-Pentamethyl-5,6,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
99 |
0.36 |
Binding ≤ 10μM
|
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2745 |
0.29 |
Binding ≤ 10μM
|
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9901 |
0.26 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9901 |
0.26 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
99 |
0.36 |
Binding ≤ 10μM
|
|
RXRB-1-E |
Retinoid X Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
RXRG-1-E |
Retinoid X Receptor Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
18 |
0.40 |
Binding ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
38 |
0.38 |
Functional ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Functional ≤ 10μM
|
|
Z80110-1-O |
CV-1 (Kidney Cells) (cluster #1 Of 2), Other |
Other |
17 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.37 |
2.81 |
-46.66 |
0 |
3 |
-1 |
53 |
362.493 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
RARB-2-E |
Retinoic Acid Receptor Beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
RARG-2-E |
Retinoic Acid Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.49 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.48 |
Binding ≤ 10μM
|
|
RXRB-2-E |
Retinoid X Receptor Beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM
|
|
RXRG-1-E |
Retinoid X Receptor Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
304 |
0.41 |
Functional ≤ 10μM
|
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
52 |
0.46 |
Functional ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
74 |
0.45 |
Functional ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
13 |
0.50 |
Functional ≤ 10μM
|
|
RXRB-1-E |
Retinoid X Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Functional ≤ 10μM
|
|
RXRG-2-E |
Retinoid X Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Functional ≤ 10μM
|
|
Z50594-7-O |
Mus Musculus (cluster #7 Of 9), Other |
Other |
58 |
0.46 |
Functional ≤ 10μM
|
|
Z80156-5-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #5 Of 12), Other |
Other |
2 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.80 |
11.58 |
-54.7 |
0 |
2 |
-1 |
40 |
299.434 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.80 |
11.01 |
-5.25 |
1 |
2 |
0 |
37 |
300.442 |
5 |
↓
|
|