|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THA-1-E |
Thyroid Hormone Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
45 |
0.43 |
Binding ≤ 10μM
|
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.91 |
8.71 |
-52 |
1 |
4 |
-1 |
70 |
327.4 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
59 |
0.36 |
Binding ≤ 10μM
|
|
TGFR1-1-E |
TGF-beta Receptor Type I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
88 |
0.35 |
Binding ≤ 10μM
|
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
88 |
0.35 |
Binding ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
152 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
9.89 |
-13.83 |
0 |
5 |
0 |
53 |
370.456 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.89 |
10.23 |
-28.84 |
1 |
5 |
1 |
54 |
371.464 |
4 |
↓
|
|
|
|
Analogs
-
38605016
-
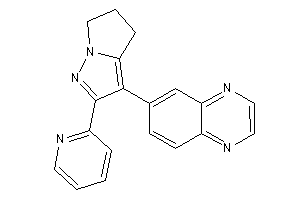
-
3959536
-
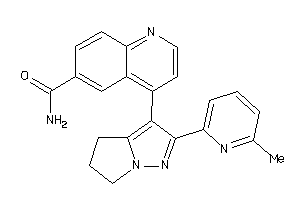
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7900 |
0.30 |
Binding ≤ 10μM
|
|
TGFR1-1-E |
TGF-beta Receptor Type I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
104 |
0.41 |
Binding ≤ 10μM
|
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
104 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
9.04 |
-13.14 |
0 |
4 |
0 |
44 |
312.376 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.42 |
9.38 |
-29.21 |
1 |
4 |
1 |
45 |
313.384 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THA-1-E |
Thyroid Hormone Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.38 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
7000 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
6.5 |
-41.98 |
4 |
5 |
0 |
97 |
650.976 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.64 |
11.6 |
-40.7 |
1 |
2 |
1 |
22 |
262.417 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.64 |
9.14 |
-5.85 |
0 |
2 |
0 |
20 |
261.409 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.98 |
10.01 |
-6.53 |
0 |
1 |
0 |
17 |
216.324 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THA-1-E |
Thyroid Hormone Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.45 |
-2.52 |
-43.51 |
1 |
4 |
-1 |
69 |
634.953 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THA-1-E |
Thyroid Hormone Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z50597-5-O |
Rattus Norvegicus (cluster #5 Of 5), Other |
Other |
1 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.93 |
-2.6 |
-43.04 |
1 |
4 |
-1 |
69 |
620.926 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THA-1-E |
Thyroid Hormone Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
THA-1-E |
Thyroid Hormone Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.55 |
Functional ≤ 10μM
|
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
1500 |
0.35 |
Binding ≤ 10μM |
|
Z80092-2-O |
CHO-K1 (Ovarian Cells) (cluster #2 Of 2), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
-5.47 |
-43.48 |
4 |
5 |
0 |
97 |
650.976 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.24 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.34 |
Binding ≤ 10μM
|
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.23 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.24 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.25 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
THA-1-E |
Thyroid Hormone Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
650 |
0.28 |
Binding ≤ 10μM
|
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.28 |
Binding ≤ 10μM
|
|
CP2C8-1-E |
Cytochrome P450 2C8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.26 |
ADME/T ≤ 10μM
|
|
LEF-2-B |
Anthrax Lethal Factor (cluster #2 Of 2), Bacterial |
Bacteria |
3500 |
0.25 |
Binding ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
62 |
0.33 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
2500 |
0.25 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
9000 |
0.23 |
Functional ≤ 10μM
|
|
Z80773-1-O |
CHRC/5 Cell Line (cluster #1 Of 1), Other |
Other |
2300 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.36 |
2.42 |
-36.05 |
1 |
4 |
1 |
43 |
646.327 |
11 |
↓
|
|