|
|
Analogs
-
2147303
-
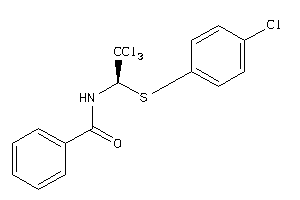
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.14 |
-0.95 |
-4.86 |
1 |
2 |
0 |
29 |
409.165 |
5 |
↓
|
|
|
|
Analogs
-
2147303
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.14 |
-0.87 |
-5.25 |
1 |
2 |
0 |
29 |
409.165 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
15 |
0.58 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
7.2 |
-7.64 |
1 |
3 |
0 |
46 |
361.077 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
15 |
0.58 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
7.26 |
-9.14 |
1 |
3 |
0 |
46 |
361.077 |
5 |
↓
|
|
|
|
Analogs
-
2147305
-
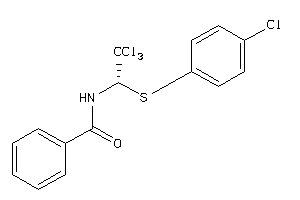
-
3138918
-
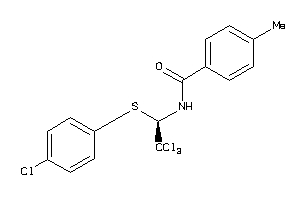
-
3138919
-
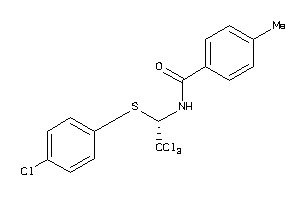
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
66 |
0.46 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.76 |
7.74 |
-4.11 |
1 |
2 |
0 |
33 |
395.138 |
5 |
↓
|
|
|
|
Analogs
-
3138918
-

-
3138919
-

-
2147303
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
66 |
0.46 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.76 |
7.66 |
-4.12 |
1 |
2 |
0 |
33 |
395.138 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
115 |
0.40 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.75 |
-1.23 |
-5.88 |
1 |
3 |
0 |
38 |
425.164 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
115 |
0.40 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.75 |
-1.16 |
-6.22 |
1 |
3 |
0 |
38 |
425.164 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.47 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.42 |
11.08 |
-7.99 |
1 |
5 |
0 |
75 |
419.717 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.47 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.42 |
11.12 |
-8.23 |
1 |
5 |
0 |
75 |
419.717 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1900 |
1.33 |
Binding ≤ 10μM |
|
TRPA1-5-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5813 |
1.22 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
3.02 |
-27.34 |
1 |
1 |
1 |
14 |
100.166 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.25 |
2.45 |
-1.96 |
0 |
1 |
0 |
12 |
99.158 |
2 |
↓
|
|
|
|
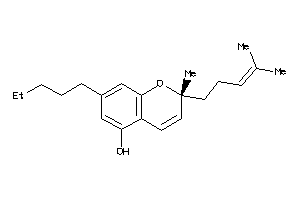
Analogs
-
1531364
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
60 |
0.44 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.50 |
1.39 |
-5.18 |
1 |
2 |
0 |
29 |
314.469 |
7 |
↓
|
|
|
|
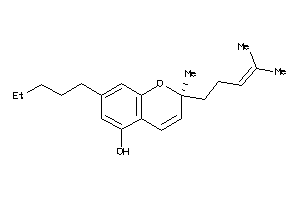
Analogs
-
1531363
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
60 |
0.44 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.50 |
1.4 |
-5.17 |
1 |
2 |
0 |
29 |
314.469 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
41 |
0.45 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
80 |
0.43 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM |
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.45 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
11 |
0.48 |
Functional ≤ 10μM
|
|
TRPA1-3-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
230 |
0.40 |
Functional ≤ 10μM |
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.69 |
1.18 |
-5.02 |
1 |
2 |
0 |
29 |
314.469 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
760 |
0.37 |
Binding ≤ 10μM |
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
880 |
0.37 |
Binding ≤ 10μM |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
700 |
0.37 |
Functional ≤ 10μM |
|
TRPM8-1-E |
Transient Receptor Potential Cation Channel Subfamily M Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.41 |
Functional ≤ 10μM |
|
TRPV1-2-E |
Vanilloid Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Functional ≤ 10μM |
|
TRPV2-1-E |
Transient Receptor Potential Cation Channel Subfamily V Member 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.35 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.84 |
0.36 |
-4.27 |
2 |
2 |
0 |
40 |
316.485 |
9 |
↓
|
|