|
|
Analogs
-
655345
-
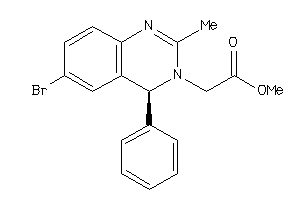
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6800 |
0.31 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
10.54 |
-12.38 |
0 |
4 |
0 |
42 |
373.25 |
4 |
↓
|
|
|
|
Analogs
-
655343
-
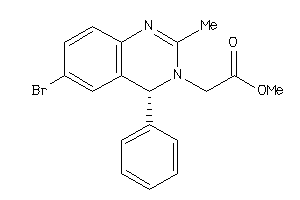
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6800 |
0.31 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
10.36 |
-9.62 |
0 |
4 |
0 |
42 |
373.25 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9400 |
0.31 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
1.82 |
-9.69 |
0 |
4 |
0 |
41 |
328.799 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9400 |
0.31 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
1.85 |
-9.57 |
0 |
4 |
0 |
41 |
328.799 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.29 |
Binding ≤ 10μM
|
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
960 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
1.74 |
-20.31 |
1 |
7 |
0 |
101 |
370.405 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.40 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
1010 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
8.37 |
-16.62 |
1 |
5 |
0 |
75 |
270.288 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
150 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
1.19 |
-16.76 |
1 |
5 |
0 |
74 |
298.342 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.37 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
820 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
8.44 |
-16.71 |
1 |
5 |
0 |
75 |
288.278 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7800 |
0.36 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
950 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
-2.04 |
-50.03 |
1 |
4 |
1 |
55 |
296.412 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.46 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
550 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.63 |
-1.36 |
-51.18 |
1 |
4 |
1 |
55 |
256.347 |
5 |
↓
|
|
|
|
Analogs
-
34543218
-
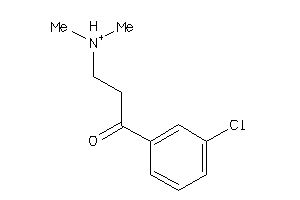
-
39372435
-
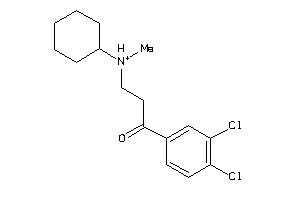
-
1572430
-
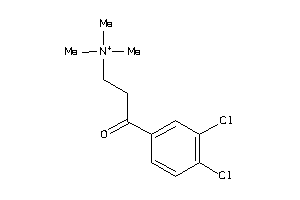
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.51 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
1000 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
1.99 |
-46.01 |
1 |
2 |
1 |
21 |
247.145 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
340 |
0.57 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
680 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
7.73 |
-54.13 |
1 |
5 |
1 |
67 |
223.252 |
5 |
↓
|
|
|
|
Analogs
-
22141174
-
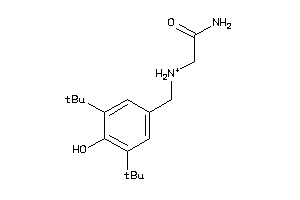
-
1533450
-
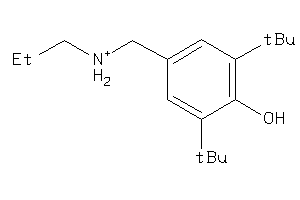
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
3010 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
-1.51 |
-38.79 |
4 |
3 |
1 |
57 |
280.432 |
6 |
↓
|
|