|
|
Analogs
-
8552123
-
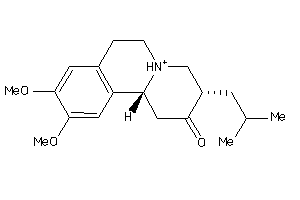
-
751
-
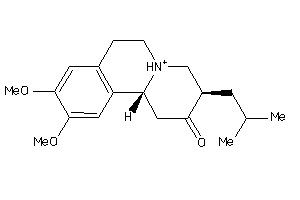
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
9.31 |
-50.64 |
1 |
4 |
1 |
40 |
318.437 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.86 |
7.34 |
-9.11 |
0 |
4 |
0 |
39 |
317.429 |
4 |
↓
|
|
|
|
Analogs
-
8552123
-

-
751
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
9.32 |
-50.72 |
1 |
4 |
1 |
40 |
318.437 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.86 |
7.18 |
-8.67 |
0 |
4 |
0 |
39 |
317.429 |
4 |
↓
|
|
|
|
Analogs
-
56446
-
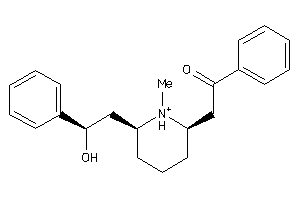
-
156831
-
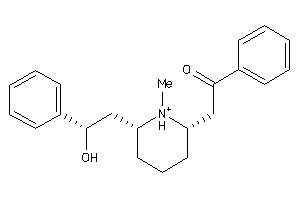
-
156931
-
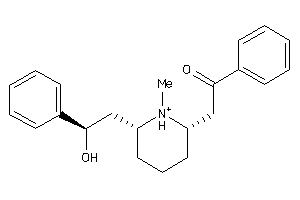
-
519487
-
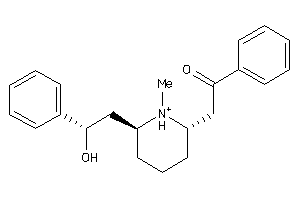
-
1081316
-
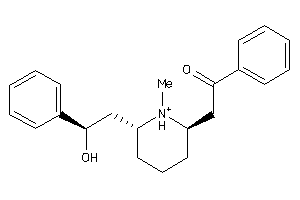
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA2-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA5-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA6-3-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6260 |
0.29 |
Binding ≤ 10μM
|
|
ACHA9-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB2-5-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB3-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB4-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2760 |
0.31 |
Binding ≤ 10μM
|
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6260 |
0.29 |
Binding ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.39 |
ADME/T ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
4 |
0.47 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
40 |
0.41 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
10.48 |
-45.25 |
2 |
3 |
1 |
42 |
338.471 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.73 |
8.25 |
-8.89 |
1 |
3 |
0 |
41 |
337.463 |
6 |
↓
|
|
|
|
Analogs
-
519487
-

-
1624
-
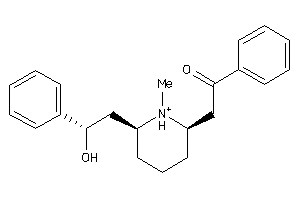
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6260 |
0.29 |
Binding ≤ 10μM
|
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2760 |
0.31 |
Binding ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.39 |
ADME/T ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
4 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
10.67 |
-44.31 |
2 |
3 |
1 |
42 |
338.471 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.73 |
8.5 |
-7.31 |
1 |
3 |
0 |
41 |
337.463 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,3R,11bR)-3-Isobutyl-9,10-dimethoxy-2,3,4,6,7,11b-hexahydro-1H-pyrido[2,1-a]isoquinolin-2-ol
(2S,3R,11bR)-3-Isobutyl-9,10-dim…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
6.96 |
-40.46 |
2 |
4 |
1 |
43 |
320.453 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.05 |
4.81 |
-7.47 |
1 |
4 |
0 |
42 |
319.445 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2460 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
6.95 |
-40.63 |
2 |
4 |
1 |
43 |
320.453 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.05 |
4.76 |
-7.46 |
1 |
4 |
0 |
42 |
319.445 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
5759845
-
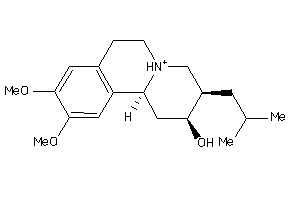
-
5760071
-
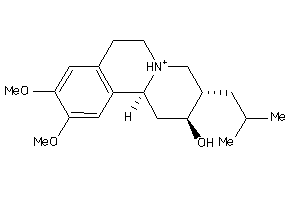
-
34325316
-
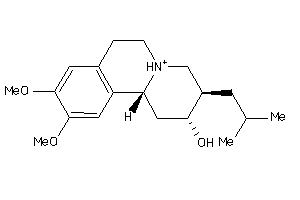
Draw
Identity
99%
90%
80%
70%
Popular Name:
2H-Benzo(a)quinolizin-2-ol, 2-ethyl-1,3,4,6,7,11b-hexahydro-3-isobutyl-9,10-dimethoxy-; 2H-Benzo(a)quinolizin-2-ol, 2-ethyl-1,3,4,6,7,11b-hexahydro-9,10-dimethoxy-3-(2-methylpropyl)-; BRN 1547216; LS-40278; Ro 4-1284; Ro 4-12884
2H-Benzo(a)quinolizin-2-ol, 2-et…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
28 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
0.65 |
-38.55 |
2 |
4 |
1 |
43 |
348.507 |
5 |
↓
|
|
|
|
Analogs
-
5759845
-

-
5760071
-

-
34325316
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
2H-Benzo(a)quinolizin-2-ol, 2-ethyl-1,3,4,6,7,11b-hexahydro-3-isobutyl-9,10-dimethoxy-; 2H-Benzo(a)quinolizin-2-ol, 2-ethyl-1,3,4,6,7,11b-hexahydro-9,10-dimethoxy-3-(2-methylpropyl)-; BRN 1547216; LS-40278; Ro 4-1284; Ro 4-12884
2H-Benzo(a)quinolizin-2-ol, 2-et…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
28 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
0.71 |
-43.17 |
2 |
4 |
1 |
43 |
348.507 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2H-Benzo(a)quinolizin-2-ol, 2-ethyl-1,3,4,6,7,11b-hexahydro-3-isobutyl-9,10-dimethoxy-; 2H-Benzo(a)quinolizin-2-ol, 2-ethyl-1,3,4,6,7,11b-hexahydro-9,10-dimethoxy-3-(2-methylpropyl)-; BRN 1547216; LS-40278; Ro 4-1284; Ro 4-12884
2H-Benzo(a)quinolizin-2-ol, 2-et…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
28 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
0.43 |
-43.83 |
2 |
4 |
1 |
43 |
348.507 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2H-Benzo(a)quinolizin-2-ol, 2-ethyl-1,3,4,6,7,11b-hexahydro-3-isobutyl-9,10-dimethoxy-; 2H-Benzo(a)quinolizin-2-ol, 2-ethyl-1,3,4,6,7,11b-hexahydro-9,10-dimethoxy-3-(2-methylpropyl)-; BRN 1547216; LS-40278; Ro 4-1284; Ro 4-12884
2H-Benzo(a)quinolizin-2-ol, 2-et…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
28 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
0.66 |
-40.85 |
2 |
4 |
1 |
43 |
348.507 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
5760071
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
2H-Benzo[a]quinolizin-2-ol, 1,3,4,6,7,11b-hexahydro-9,10-dimethoxy-3-(2-methylpropyl)-, (2R,3R,11bR)-
2H-Benzo[a]quinolizin-2-ol, 1,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
6.79 |
-44.35 |
2 |
4 |
1 |
43 |
320.453 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.05 |
4.64 |
-7.9 |
1 |
4 |
0 |
42 |
319.445 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
5714404
-
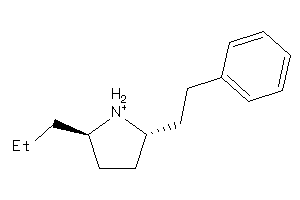
-
15776789
-
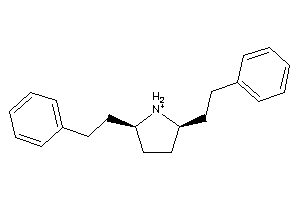
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3060 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.12 |
0.66 |
-44.08 |
2 |
1 |
1 |
16 |
280.435 |
6 |
↓
|
|
|
|
Analogs
-
5714404
-

-
15776789
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
54 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.12 |
12.34 |
-43.14 |
2 |
1 |
1 |
17 |
280.435 |
6 |
↓
|
|
|
|
|