|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100751-1-O |
PMNL (Polymorphonuclear Leukocytes) (cluster #1 Of 2), Other |
Other |
6000 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
4.16 |
-17 |
3 |
8 |
0 |
130 |
374.345 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.55 |
5.73 |
-110.55 |
1 |
8 |
-2 |
136 |
372.329 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.55 |
4.95 |
-46.6 |
2 |
8 |
-1 |
133 |
373.337 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100751-1-O |
PMNL (Polymorphonuclear Leukocytes) (cluster #1 Of 2), Other |
Other |
8000 |
0.26 |
Functional ≤ 10μM
|
|
Z80901-1-O |
HaCaT (Keratinocytes) (cluster #1 Of 2), Other |
Other |
2600 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
7.88 |
-57.17 |
1 |
7 |
-1 |
105 |
373.381 |
6 |
↓
|
|
|
|
Analogs
-
32148112
-
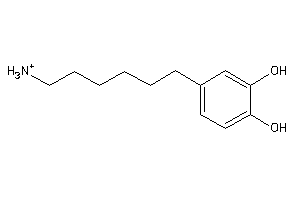
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.51 |
Binding ≤ 10μM
|
|
IGF1R-1-E |
Insulin-like Growth Factor 1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.38 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
180 |
0.43 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.35 |
Binding ≤ 10μM
|
|
LOX12-1-E |
Arachidonate 12-lipoxygenase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5100 |
0.34 |
Binding ≤ 10μM
|
|
LOX12-1-E |
Arachidonate 12-lipoxygenase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.33 |
Binding ≤ 10μM
|
|
LOX15-1-E |
Arachidonate 15-lipoxygenase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
110 |
0.44 |
Binding ≤ 10μM
|
|
LOX15-1-E |
Arachidonate 15-lipoxygenase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5100 |
0.34 |
Binding ≤ 10μM |
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5000 |
0.34 |
Binding ≤ 10μM
|
|
LT4R1-1-E |
Leukotriene B4 Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
85 |
0.45 |
Binding ≤ 10μM
|
|
Q9BEG3-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
LOX5-6-E |
Arachidonate 5-lipoxygenase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
5000 |
0.34 |
Functional ≤ 10μM
|
|
Q9BEG3-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Functional ≤ 10μM
|
|
Z100274-1-O |
Liposome (cluster #1 Of 1), Other |
Other |
2000 |
0.36 |
Functional ≤ 10μM
|
|
Z100751-1-O |
PMNL (Polymorphonuclear Leukocytes) (cluster #1 Of 2), Other |
Other |
400 |
0.41 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
7943 |
0.32 |
Functional ≤ 10μM
|
|
Z50587-3-O |
Homo Sapiens (cluster #3 Of 9), Other |
Other |
8000 |
0.32 |
Functional ≤ 10μM |
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
5000 |
0.34 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
2300 |
0.36 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
8500 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
2.05 |
-11.23 |
4 |
4 |
0 |
81 |
302.37 |
5 |
↓
|
|
|
|
Analogs
-
32148112
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.51 |
Binding ≤ 10μM
|
|
IGF1R-1-E |
Insulin-like Growth Factor 1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.38 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.35 |
Binding ≤ 10μM
|
|
LOX12-1-E |
Arachidonate 12-lipoxygenase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.33 |
Binding ≤ 10μM
|
|
LOX15-1-E |
Arachidonate 15-lipoxygenase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5100 |
0.34 |
Binding ≤ 10μM |
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5000 |
0.34 |
Binding ≤ 10μM
|
|
LT4R1-1-E |
Leukotriene B4 Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
85 |
0.45 |
Binding ≤ 10μM
|
|
Q9BEG3-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
LOX5-6-E |
Arachidonate 5-lipoxygenase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
5000 |
0.34 |
Functional ≤ 10μM
|
|
Q9BEG3-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Functional ≤ 10μM
|
|
Z100274-1-O |
Liposome (cluster #1 Of 1), Other |
Other |
2000 |
0.36 |
Functional ≤ 10μM
|
|
Z100751-1-O |
PMNL (Polymorphonuclear Leukocytes) (cluster #1 Of 2), Other |
Other |
400 |
0.41 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
7943 |
0.32 |
Functional ≤ 10μM
|
|
Z50587-3-O |
Homo Sapiens (cluster #3 Of 9), Other |
Other |
8000 |
0.32 |
Functional ≤ 10μM |
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
5000 |
0.34 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
2300 |
0.36 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
8500 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
2.06 |
-11.4 |
4 |
4 |
0 |
81 |
302.37 |
5 |
↓
|
|
|
|
Analogs
-
32148112
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.51 |
Binding ≤ 10μM
|
|
IGF1R-1-E |
Insulin-like Growth Factor 1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.38 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
180 |
0.43 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.35 |
Binding ≤ 10μM
|
|
LOX12-1-E |
Arachidonate 12-lipoxygenase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5100 |
0.34 |
Binding ≤ 10μM
|
|
LOX12-1-E |
Arachidonate 12-lipoxygenase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.33 |
Binding ≤ 10μM
|
|
LOX15-1-E |
Arachidonate 15-lipoxygenase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
110 |
0.44 |
Binding ≤ 10μM
|
|
LOX15-1-E |
Arachidonate 15-lipoxygenase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5100 |
0.34 |
Binding ≤ 10μM |
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5000 |
0.34 |
Binding ≤ 10μM
|
|
LT4R1-1-E |
Leukotriene B4 Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
85 |
0.45 |
Binding ≤ 10μM
|
|
Q9BEG3-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
LOX5-6-E |
Arachidonate 5-lipoxygenase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
5000 |
0.34 |
Functional ≤ 10μM
|
|
Q9BEG3-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Functional ≤ 10μM
|
|
Z100274-1-O |
Liposome (cluster #1 Of 1), Other |
Other |
2000 |
0.36 |
Functional ≤ 10μM
|
|
Z100751-1-O |
PMNL (Polymorphonuclear Leukocytes) (cluster #1 Of 2), Other |
Other |
400 |
0.41 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
7943 |
0.32 |
Functional ≤ 10μM
|
|
Z50587-3-O |
Homo Sapiens (cluster #3 Of 9), Other |
Other |
8000 |
0.32 |
Functional ≤ 10μM |
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
5000 |
0.34 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
2300 |
0.36 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
8500 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
2.04 |
-11.3 |
4 |
4 |
0 |
81 |
302.37 |
5 |
↓
|
|