|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 6), Bacterial |
Bacteria |
6 |
0.46 |
Binding ≤ 10μM
|
|
CP51-1-B |
Sterol 14-alpha Demethylase (cluster #1 Of 2), Bacterial |
Bacteria |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
82 |
0.40 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.39 |
Binding ≤ 10μM
|
|
KCNA3-1-E |
Voltage-gated Potassium Channel Subunit Kv1.3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.29 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3020 |
0.31 |
Binding ≤ 10μM
|
|
KCNN4-1-E |
Intermediate Conductance Calcium-activated Potassium Channel Protein 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.40 |
Binding ≤ 10μM
|
|
MDHM-1-E |
Malate Dehydrogenase, Mitochondrial (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6700 |
0.29 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4800 |
0.30 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.41 |
ADME/T ≤ 10μM
|
|
Q96W81-1-F |
14-alpha Sterol Demethylase (cluster #1 Of 1), Fungal |
Fungi |
103 |
0.39 |
Binding ≤ 10μM
|
|
Q4WNT5-1-O |
14-alpha Sterol Demethylase Cyp51A (cluster #1 Of 1), Other |
Other |
4790 |
0.30 |
Binding ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
6540 |
0.29 |
Functional ≤ 10μM
|
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
60 |
0.40 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
250 |
0.37 |
Functional ≤ 10μM
|
|
Z80682-8-O |
A549 (Lung Carcinoma Cells) (cluster #8 Of 11), Other |
Other |
5100 |
0.30 |
Functional ≤ 10μM
|
|
Z80951-2-O |
NIH3T3 (Fibroblasts) (cluster #2 Of 4), Other |
Other |
630 |
0.35 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
3000 |
0.31 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.47 |
13.43 |
-5.78 |
0 |
2 |
0 |
18 |
344.845 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.47 |
13.99 |
-28.18 |
1 |
2 |
1 |
19 |
345.853 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1510 |
0.34 |
ADME/T ≤ 10μM |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
9 |
0.47 |
Functional ≤ 10μM |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
27 |
0.44 |
Functional ≤ 10μM |
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
9 |
0.47 |
Functional ≤ 10μM |
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
21 |
0.45 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
5.64 |
-9.04 |
3 |
4 |
0 |
57 |
337.81 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 99 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.20 |
8.91 |
-32.71 |
0 |
5 |
1 |
41 |
336.367 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.40 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z100501-1-O |
Lu1 (Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
80 |
0.52 |
Functional ≤ 10μM
|
|
Z103204-1-O |
A427 (cluster #1 Of 4), Other |
Other |
890 |
0.45 |
Functional ≤ 10μM
|
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
470 |
0.47 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
501 |
0.46 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
73 |
0.53 |
Functional ≤ 10μM
|
|
Z80008-2-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
890 |
0.45 |
Functional ≤ 10μM
|
|
Z80036-2-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #2 Of 3), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80145-1-O |
H69 (cluster #1 Of 4), Other |
Other |
1800 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-10-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #10 Of 12), Other |
Other |
660 |
0.46 |
Functional ≤ 10μM
|
|
Z80164-3-O |
HT-1080 (Fibrosarcoma Cells) (cluster #3 Of 6), Other |
Other |
4300 |
0.40 |
Functional ≤ 10μM
|
|
Z80186-10-O |
K562 (Erythroleukemia Cells) (cluster #10 Of 11), Other |
Other |
380 |
0.47 |
Functional ≤ 10μM
|
|
Z80193-4-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #4 Of 12), Other |
Other |
500 |
0.46 |
Functional ≤ 10μM |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
430 |
0.47 |
Functional ≤ 10μM
|
|
Z80322-1-O |
NCI-H358 (Lung Carcinama Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80509-1-O |
SNU-638 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.42 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
800 |
0.45 |
Functional ≤ 10μM
|
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
470 |
0.47 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
410 |
0.47 |
Functional ≤ 10μM
|
|
Z81024-8-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #8 Of 8), Other |
Other |
510 |
0.46 |
Functional ≤ 10μM
|
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 6), Other |
Other |
812 |
0.45 |
Functional ≤ 10μM
|
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 6), Other |
Other |
1780 |
0.42 |
Functional ≤ 10μM |
|
Z81170-5-O |
LNCaP (Prostate Carcinoma) (cluster #5 Of 5), Other |
Other |
3250 |
0.40 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
310 |
0.48 |
Functional ≤ 10μM
|
|
Z81252-10-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #10 Of 11), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
|
Z81277-1-O |
NCI-N417 (cluster #1 Of 2), Other |
Other |
870 |
0.45 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 4), Other |
Other |
2600 |
0.41 |
ADME/T ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 1), Other |
Other |
5010 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
190 |
0.50 |
ADME/T ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
3820 |
0.40 |
ADME/T ≤ 10μM |
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
5500 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
2070 |
0.42 |
ADME/T ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1820 |
0.42 |
ADME/T ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 1), Other |
Other |
3 |
0.63 |
ADME/T ≤ 10μM
|
|
Z81057-4-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #4 Of 5), Other |
Other |
370 |
0.47 |
ADME/T ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
5300 |
0.39 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.28 |
8.05 |
-7.84 |
1 |
2 |
0 |
29 |
246.313 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.28 |
8.52 |
-35.14 |
2 |
2 |
1 |
30 |
247.321 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
6.35 |
-85.33 |
5 |
4 |
2 |
63 |
329.831 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
5.92 |
-48.44 |
4 |
4 |
1 |
62 |
328.823 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
6800 |
0.33 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
-1.86 |
-12.24 |
2 |
4 |
0 |
54 |
311.772 |
2 |
↓
|
|
|
|
Analogs
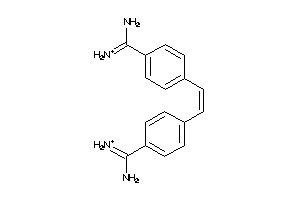
-
4777954
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
27 |
0.53 |
Functional ≤ 10μM
|
|
Z50461-2-O |
Leishmania Mexicana (cluster #2 Of 3), Other |
Other |
9000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
-3.82 |
-72.78 |
8 |
4 |
2 |
103 |
266.348 |
4 |
↓
|
|
|
|
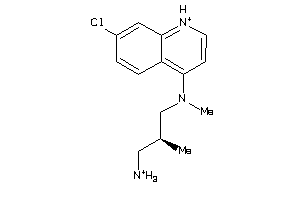
Analogs
-
42455656
-
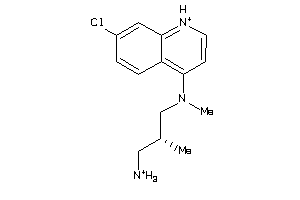
-
42455657
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7 |
0.57 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
23 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
-0.33 |
-85.24 |
3 |
3 |
2 |
30 |
293.842 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-4-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #4 Of 9), Other |
Other |
868 |
0.35 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.58 |
9.62 |
-11.58 |
0 |
4 |
0 |
46 |
318.376 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
51 |
0.41 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
62 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.22 |
2.82 |
-85.32 |
3 |
3 |
2 |
30 |
355.448 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.96 |
11.84 |
-46.11 |
0 |
3 |
-1 |
57 |
365.836 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
5.55 |
9.3 |
-8.83 |
2 |
3 |
0 |
58 |
366.844 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.55 |
10.22 |
-45.56 |
1 |
3 |
-1 |
60 |
365.836 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
910 |
0.42 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
13 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
8.73 |
-94.78 |
3 |
3 |
2 |
31 |
291.826 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.62 |
6.06 |
-7.06 |
1 |
3 |
0 |
28 |
289.81 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3900 |
0.30 |
Binding ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.29 |
ADME/T ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
6 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.29 |
8.78 |
-78.63 |
4 |
4 |
2 |
51 |
357.885 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.29 |
9.02 |
-33.48 |
2 |
4 |
0 |
52 |
355.869 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.48 |
7.13 |
-80.66 |
5 |
4 |
2 |
63 |
357.885 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.48 |
7.46 |
-32.55 |
3 |
4 |
0 |
65 |
355.869 |
5 |
↓
|
|
|
|
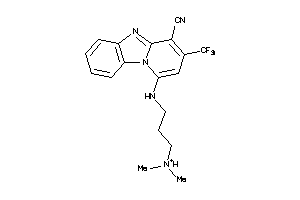
Analogs
-
4115356
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
2080 |
0.28 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.37 |
12.26 |
-61.07 |
2 |
5 |
1 |
58 |
390.433 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
240 |
0.46 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
240 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
3.99 |
-33.91 |
2 |
4 |
1 |
39 |
292.79 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.56 |
3.6 |
-8.85 |
1 |
4 |
0 |
37 |
291.782 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.56 |
6.32 |
-98.57 |
3 |
4 |
2 |
40 |
293.798 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
1338 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
-3.59 |
-89.89 |
5 |
3 |
2 |
53 |
265.788 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
1338 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
-3.58 |
-89.86 |
5 |
3 |
2 |
53 |
265.788 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
5300 |
0.24 |
Functional ≤ 10μM |
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
3000 |
0.25 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.30 |
-3.39 |
-24.54 |
5 |
10 |
0 |
163 |
434.397 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
2050 |
0.35 |
Functional ≤ 10μM |
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 2), Other |
Other |
10000 |
0.30 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
-1.96 |
-11.32 |
2 |
4 |
0 |
54 |
346.217 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
690 |
0.39 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
7.02 |
-10.6 |
2 |
4 |
0 |
54 |
295.317 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
4720 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
6.99 |
-12.74 |
2 |
4 |
0 |
54 |
295.317 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-4-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #4 Of 9), Other |
Other |
95 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
-2.73 |
-14.98 |
5 |
9 |
0 |
162 |
318.193 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.88 |
-1.68 |
-48.41 |
4 |
9 |
-1 |
164 |
317.185 |
0 |
↓
|
|
|
|
Analogs
-
1571100
-
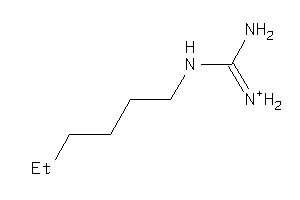
Draw
Identity
99%
90%
80%
70%
Popular Name:
EINECS 230-888-7; Guanidine, N,N'''-1,6-hexanediylbis-, dihydrochloride; Guanidine, hexanedi-, dihydrochloride; Hexamethylenediguanidine dihydrochloride; Hexanediguanidine dihydrochloride; LS-73701; N,N'''-1,6-Hexanediylbisguanidine dihydrochloride; NSC 3
EINECS 230-888-7; Guanidine, N,N…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-5-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #5 Of 9), Other |
Other |
445 |
0.64 |
Functional ≤ 10μM
|
|
Z50725-5-O |
Trypanosoma Brucei Rhodesiense (cluster #5 Of 7), Other |
Other |
8050 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.41 |
2.46 |
-70.53 |
10 |
6 |
2 |
127 |
202.306 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 101 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
610 |
0.36 |
ADME/T ≤ 10μM
|
|
CP2DI-1-E |
Cytochrome P450 2D18 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.34 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
94 |
0.41 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
652 |
0.36 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
81 |
0.41 |
Functional ≤ 10μM
|
|
Z50460-2-O |
Leishmania Major (cluster #2 Of 4), Other |
Other |
9530 |
0.29 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
10000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
7.18 |
-40.05 |
2 |
4 |
1 |
47 |
325.432 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.06 |
7.62 |
-89.58 |
3 |
4 |
2 |
48 |
326.44 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
9800 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
5.92 |
-27 |
2 |
2 |
1 |
30 |
183.234 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.81 |
5.53 |
-10.38 |
1 |
2 |
0 |
29 |
182.226 |
0 |
↓
|
|
|
|
Analogs
-
1733340
-
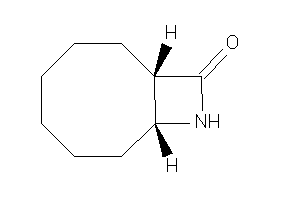
-
1733341
-
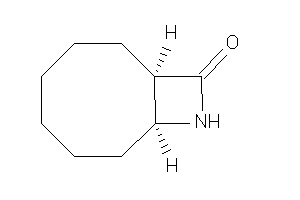
-
1733342
-
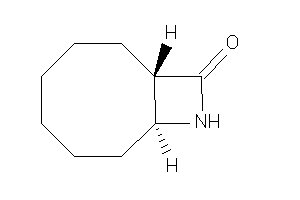
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-8-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #8 Of 9), Other |
Other |
8400 |
0.65 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
-2.61 |
-5.95 |
1 |
2 |
0 |
29 |
153.225 |
0 |
↓
|
|
|
|
Analogs
-
1733341
-

-
1733342
-

-
1733339
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-8-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #8 Of 9), Other |
Other |
8400 |
0.65 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
-2.45 |
-6.21 |
1 |
2 |
0 |
29 |
153.225 |
0 |
↓
|
|
|
|
Analogs
-
1733342
-

-
1733339
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-8-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #8 Of 9), Other |
Other |
8400 |
0.65 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
-2.24 |
-6.74 |
1 |
2 |
0 |
29 |
153.225 |
0 |
↓
|
|
|
|
Analogs
-
1733339
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-8-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #8 Of 9), Other |
Other |
8400 |
0.65 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
-3.04 |
-5.84 |
1 |
2 |
0 |
29 |
153.225 |
0 |
↓
|
|
|
|
Analogs
-
4674079
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
3160 |
0.30 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
3.55 |
-62.28 |
2 |
5 |
1 |
57 |
362.379 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
23 |
0.33 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.42 |
13.28 |
-15.85 |
0 |
5 |
0 |
48 |
427.527 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.42 |
16.02 |
-106.8 |
2 |
5 |
2 |
50 |
429.543 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
3430 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
3.42 |
-65.75 |
2 |
5 |
1 |
57 |
348.352 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
1900 |
0.30 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
3.53 |
-64.61 |
2 |
5 |
1 |
57 |
376.406 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
267 |
0.33 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
267 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.08 |
1.32 |
-84.98 |
3 |
4 |
2 |
39 |
379.548 |
10 |
↓
|
|
|
|
Analogs
-
4475165
-
-
13493525
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
16 |
0.44 |
Binding ≤ 10μM
|
|
CATC-1-E |
Dipeptidyl Peptidase I (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
770 |
0.34 |
Binding ≤ 10μM
|
|
CATH-1-E |
Cathepsin H (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
PAPA1-2-E |
Papain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.41 |
Binding ≤ 10μM
|
|
Q9NBA7-1-E |
Cysteine Protease Falcipain-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
75 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
9300 |
0.28 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
2500 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.37 |
1.58 |
-72.44 |
7 |
10 |
0 |
174 |
357.411 |
12 |
↓
|
|
|
|
Analogs
-
13493525
-

-
4095645
-
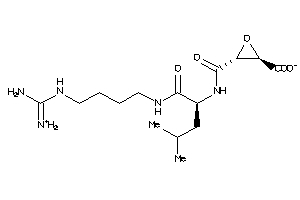
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
16 |
0.44 |
Binding ≤ 10μM
|
|
CATC-1-E |
Dipeptidyl Peptidase I (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
770 |
0.34 |
Binding ≤ 10μM
|
|
CATH-1-E |
Cathepsin H (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
PAPA1-2-E |
Papain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.41 |
Binding ≤ 10μM
|
|
Q9NBA7-1-E |
Cysteine Protease Falcipain-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
75 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
9300 |
0.28 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
2500 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.37 |
1.94 |
-73.1 |
7 |
10 |
0 |
174 |
357.411 |
12 |
↓
|
|
|
|
|
|
|
Analogs
-
6815
-
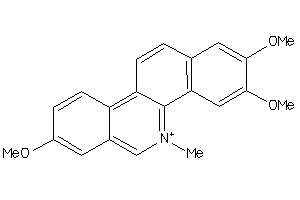
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.00 |
0.77 |
-36.44 |
1 |
5 |
1 |
51 |
350.394 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DXR-2-B |
1-deoxyxylulose-5-phosphate Reductoisomerase (cluster #2 Of 2), Bacterial |
Bacteria |
21 |
0.98 |
Binding ≤ 10μM
|
|
Z50212-4-O |
Escherichia Coli (cluster #4 Of 7), Other |
Other |
220 |
0.85 |
Functional ≤ 10μM
|
|
Z50425-5-O |
Plasmodium Falciparum (cluster #5 Of 22), Other |
Other |
480 |
0.80 |
Functional ≤ 10μM
|
|
Z50426-2-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #2 Of 9), Other |
Other |
3700 |
0.69 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.58 |
-1.63 |
-45.27 |
2 |
6 |
-1 |
101 |
182.092 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DXR-2-B |
1-deoxyxylulose-5-phosphate Reductoisomerase (cluster #2 Of 2), Bacterial |
Bacteria |
160 |
0.79 |
Binding ≤ 10μM |
|
Z50425-5-O |
Plasmodium Falciparum (cluster #5 Of 22), Other |
Other |
430 |
0.74 |
Functional ≤ 10μM
|
|
Z50426-2-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #2 Of 9), Other |
Other |
420 |
0.74 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.60 |
-2.13 |
-48.67 |
2 |
6 |
-1 |
101 |
196.119 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7 |
0.37 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
5 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
16.58 |
-43.15 |
1 |
3 |
1 |
22 |
428.987 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.72 |
17.06 |
-91.23 |
2 |
3 |
2 |
24 |
429.995 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7 |
0.37 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
5 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
16.55 |
-44.8 |
1 |
3 |
1 |
22 |
428.987 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.72 |
17.03 |
-88.61 |
2 |
3 |
2 |
24 |
429.995 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.72 |
3.89 |
-75.76 |
8 |
6 |
2 |
122 |
314.389 |
8 |
↓
|
|
|
|
Analogs
-
1758871
-
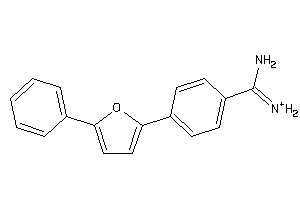
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.38 |
Binding ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.38 |
Binding ≤ 10μM
|
|
Z50420-4-O |
Trypanosoma Brucei Brucei (cluster #4 Of 7), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
16 |
0.47 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
16 |
0.47 |
Functional ≤ 10μM
|
|
Z50459-1-O |
Leishmania Donovani (cluster #1 Of 8), Other |
Other |
6400 |
0.32 |
Functional ≤ 10μM
|
|
Z50466-7-O |
Trypanosoma Cruzi (cluster #7 Of 8), Other |
Other |
6400 |
0.32 |
Functional ≤ 10μM
|
|
Z50468-4-O |
Giardia Intestinalis (cluster #4 Of 4), Other |
Other |
440 |
0.39 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z50725-1-O |
Trypanosoma Brucei Rhodesiense (cluster #1 Of 7), Other |
Other |
4500 |
0.33 |
Functional ≤ 10μM
|
|
Z80035-5-O |
B16 (Melanoma Cells) (cluster #5 Of 7), Other |
Other |
9200 |
0.31 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
6400 |
0.32 |
Functional ≤ 10μM
|
|
Z81135-1-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #1 Of 6), Other |
Other |
6700 |
0.31 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
5.7 |
-76.45 |
8 |
5 |
2 |
116 |
306.369 |
4 |
↓
|
|
|
|
Analogs
-
15449189
-
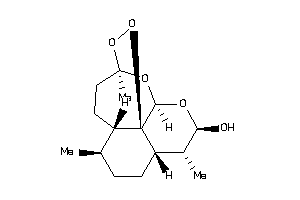
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
56 |
0.51 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
6 |
0.58 |
Functional ≤ 10μM
|
|
Z50436-3-O |
Filobasidiella Neoformans (cluster #3 Of 3), Other |
Other |
280 |
0.46 |
Functional ≤ 10μM
|
|
Z50442-2-O |
Candida Albicans (cluster #2 Of 4), Other |
Other |
880 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 12), Other |
Other |
2900 |
0.39 |
Functional ≤ 10μM
|
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
750 |
0.43 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
850 |
0.42 |
Functional ≤ 10μM
|
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
540 |
0.44 |
Functional ≤ 10μM
|
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
630 |
0.43 |
Functional ≤ 10μM
|
|
Z81057-2-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #2 Of 4), Other |
Other |
8910 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
650 |
0.43 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
460 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
3.29 |
-6.48 |
1 |
5 |
0 |
57 |
284.352 |
0 |
↓
|
|