|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
3.26 |
-51.83 |
0 |
5 |
-1 |
74 |
365.331 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AL5AP-1-E |
5-lipoxygenase Activating Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.33 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
90 |
0.31 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1100 |
0.26 |
Binding ≤ 10μM
|
|
PGES2-1-E |
Prostaglandin E Synthase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.25 |
Binding ≤ 10μM
|
|
PTGES-1-E |
Prostaglandin E Synthase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.24 |
Binding ≤ 10μM
|
|
AL5AP-1-E |
5-lipoxygenase Activating Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.28 |
Functional ≤ 10μM |
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Functional ≤ 10μM
|
|
THAS-1-E |
Thromboxane-A Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.25 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 1), Other |
Other |
3 |
0.37 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.23 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
50 |
0.32 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
200 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.31 |
17.19 |
-45.93 |
0 |
3 |
-1 |
45 |
471.086 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.24 |
Binding ≤ 10μM |
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.24 |
Binding ≤ 10μM |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Functional ≤ 10μM |
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Functional ≤ 10μM |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 1), Other |
Other |
6000 |
0.24 |
Binding ≤ 10μM |
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
10000 |
0.23 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.79 |
0.21 |
-176.06 |
11 |
14 |
0 |
259 |
445.477 |
15 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.79 |
0.11 |
-139.7 |
10 |
14 |
-1 |
257 |
444.469 |
15 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-4.79 |
-1.54 |
-133.75 |
12 |
14 |
1 |
256 |
446.485 |
15 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.86 |
4.16 |
-46.86 |
5 |
10 |
1 |
143 |
421.474 |
10 |
↓
|
|
Ref
Reference (pH 7)
|
-0.99 |
4.3 |
-19.23 |
4 |
10 |
0 |
144 |
420.466 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.86 |
5.25 |
-32.88 |
4 |
10 |
0 |
146 |
420.466 |
10 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.50 |
-5.37 |
-56.05 |
6 |
9 |
1 |
139 |
362.41 |
8 |
↓
|
|
|
|
Analogs
-
37015742
-
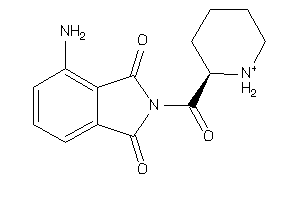
-
37015778
-
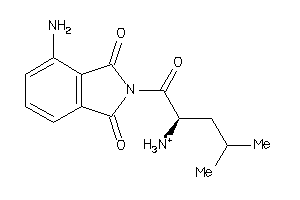
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 1), Other |
Other |
480 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.78 |
1.16 |
-21.16 |
3 |
6 |
0 |
93 |
259.265 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.32 |
4.71 |
-16.55 |
4 |
10 |
0 |
140 |
547.674 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACV1B-1-E |
Activin Receptor Type-1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
ADCK4-1-E |
Uncharacterized AarF Domain-containing Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.29 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.33 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.31 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.38 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1AL-1-E |
Casein Kinase I Isoform Alpha-like (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.39 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.35 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.34 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.34 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
M4K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.28 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
95 |
0.36 |
Binding ≤ 10μM
|
|
MK10-2-E |
C-Jun N-terminal Kinase 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.37 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.37 |
Binding ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.37 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.26 |
Binding ≤ 10μM
|
|
MRCKA-1-E |
Serine/threonine-protein Kinase MRCK-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.27 |
Binding ≤ 10μM
|
|
MRCKB-1-E |
Serine/threonine-protein Kinase MRCK Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Binding ≤ 10μM
|
|
MRCKG-1-E |
Serine/threonine-protein Kinase MRCK Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.33 |
Binding ≤ 10μM
|
|
NLK-1-E |
Serine/threonine Protein Kinase NLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.39 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.30 |
Binding ≤ 10μM
|
|
PTK6-1-E |
Tyrosine-protein Kinase BRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
Q5SC61-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.31 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.40 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4600 |
0.28 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5300 |
0.27 |
Binding ≤ 10μM
|
|
ST32B-1-E |
Serine/threonine-protein Kinase 32B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.29 |
Binding ≤ 10μM
|
|
STK36-1-E |
Serine/threonine-protein Kinase 36 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.31 |
Binding ≤ 10μM
|
|
TGFR1-1-E |
TGF-beta Receptor Type I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7100 |
0.27 |
Binding ≤ 10μM
|
|
TGFR2-1-E |
TGF-beta Receptor Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.30 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.28 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.32 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.29 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.28 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
80 |
0.37 |
Functional ≤ 10μM |
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
TNFA-1-E |
TNF-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
80 |
0.37 |
Functional ≤ 10μM |
|
Z102116-1-O |
Toxoplasma Gondii RH (cluster #1 Of 2), Other |
Other |
8500 |
0.26 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
2500 |
0.29 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
940 |
0.31 |
Functional ≤ 10μM
|
|
Z80523-1-O |
SW1353 (cluster #1 Of 2), Other |
Other |
50 |
0.38 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
72 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
8.8 |
-21.46 |
1 |
4 |
0 |
59 |
377.444 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.10 |
9.25 |
-46.75 |
2 |
4 |
1 |
60 |
378.452 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.10 |
9.24 |
-46.38 |
2 |
4 |
1 |
60 |
378.452 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACV1B-1-E |
Activin Receptor Type-1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
ADCK4-1-E |
Uncharacterized AarF Domain-containing Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.29 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.33 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.31 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.38 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1AL-1-E |
Casein Kinase I Isoform Alpha-like (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.39 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.35 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.34 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.34 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
M4K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.28 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
95 |
0.36 |
Binding ≤ 10μM
|
|
MK10-2-E |
C-Jun N-terminal Kinase 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.37 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.37 |
Binding ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.37 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.26 |
Binding ≤ 10μM
|
|
MRCKA-1-E |
Serine/threonine-protein Kinase MRCK-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.27 |
Binding ≤ 10μM
|
|
MRCKB-1-E |
Serine/threonine-protein Kinase MRCK Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Binding ≤ 10μM
|
|
MRCKG-1-E |
Serine/threonine-protein Kinase MRCK Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.33 |
Binding ≤ 10μM
|
|
NLK-1-E |
Serine/threonine Protein Kinase NLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.39 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.30 |
Binding ≤ 10μM
|
|
PTK6-1-E |
Tyrosine-protein Kinase BRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
Q5SC61-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.31 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.40 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4600 |
0.28 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5300 |
0.27 |
Binding ≤ 10μM
|
|
ST32B-1-E |
Serine/threonine-protein Kinase 32B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.29 |
Binding ≤ 10μM
|
|
STK36-1-E |
Serine/threonine-protein Kinase 36 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.31 |
Binding ≤ 10μM
|
|
TGFR1-1-E |
TGF-beta Receptor Type I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7100 |
0.27 |
Binding ≤ 10μM
|
|
TGFR2-1-E |
TGF-beta Receptor Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.30 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.28 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.32 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.29 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.28 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
80 |
0.37 |
Functional ≤ 10μM |
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
TNFA-1-E |
TNF-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
80 |
0.37 |
Functional ≤ 10μM |
|
Z102116-1-O |
Toxoplasma Gondii RH (cluster #1 Of 2), Other |
Other |
8500 |
0.26 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
2500 |
0.29 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
940 |
0.31 |
Functional ≤ 10μM
|
|
Z80523-1-O |
SW1353 (cluster #1 Of 2), Other |
Other |
50 |
0.38 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
72 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
8.81 |
-16.64 |
1 |
4 |
0 |
59 |
377.444 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.10 |
8.8 |
-16.98 |
1 |
4 |
0 |
59 |
377.444 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.10 |
9.25 |
-46.22 |
2 |
4 |
1 |
60 |
378.452 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
6032 |
0.25 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
9603 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.50 |
9.8 |
-10.06 |
1 |
4 |
0 |
47 |
433.739 |
5 |
↓
|
|
|
|
Analogs
-
14556041
-
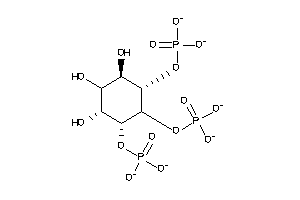
-
25757731
-
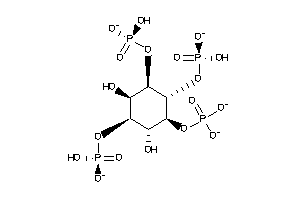
-
43308011
-
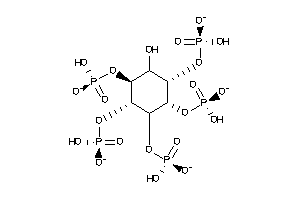
-
12494830
-
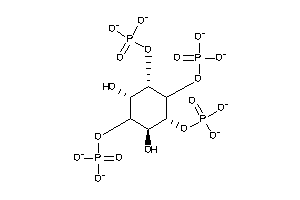
-
12494942
-
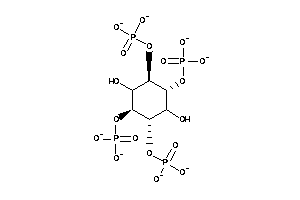
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITPR1-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
ITPR2-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
ITPR3-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
Z50587-2-O |
Homo Sapiens (cluster #2 Of 9), Other |
Other |
3900 |
0.27 |
Functional ≤ 10μM |
|
Z50592-2-O |
Oryctolagus Cuniculus (cluster #2 Of 8), Other |
Other |
9670 |
0.25 |
Functional ≤ 10μM |
|
Z80471-1-O |
SH-SY5 (Bone Marrow Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
77 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.07 |
-9.46 |
-434.04 |
5 |
18 |
-5 |
322 |
495.032 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.07 |
-6.03 |
-993.35 |
2 |
18 |
-8 |
330 |
492.008 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.07 |
-7.17 |
-794.92 |
3 |
18 |
-7 |
327 |
493.016 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITPR1-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
ITPR2-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
ITPR3-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
Z50587-2-O |
Homo Sapiens (cluster #2 Of 9), Other |
Other |
3900 |
0.27 |
Functional ≤ 10μM |
|
Z50592-2-O |
Oryctolagus Cuniculus (cluster #2 Of 8), Other |
Other |
9670 |
0.25 |
Functional ≤ 10μM |
|
Z80471-1-O |
SH-SY5 (Bone Marrow Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
77 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.07 |
-9.59 |
-433.57 |
5 |
18 |
-5 |
322 |
495.032 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.07 |
-7.3 |
-786.57 |
3 |
18 |
-7 |
327 |
493.016 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.07 |
-6.16 |
-991.09 |
2 |
18 |
-8 |
330 |
492.008 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITPR1-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
ITPR2-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
ITPR3-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
Z50587-2-O |
Homo Sapiens (cluster #2 Of 9), Other |
Other |
3900 |
0.27 |
Functional ≤ 10μM |
|
Z50592-2-O |
Oryctolagus Cuniculus (cluster #2 Of 8), Other |
Other |
9670 |
0.25 |
Functional ≤ 10μM |
|
Z80471-1-O |
SH-SY5 (Bone Marrow Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
77 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.07 |
-9.91 |
-438.18 |
5 |
18 |
-5 |
322 |
495.032 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.07 |
-7.59 |
-789.92 |
3 |
18 |
-7 |
327 |
493.016 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-5.07 |
-11.06 |
-288.63 |
6 |
18 |
-4 |
319 |
496.04 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITPR1-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
ITPR2-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
ITPR3-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
Z50587-2-O |
Homo Sapiens (cluster #2 Of 9), Other |
Other |
3900 |
0.27 |
Functional ≤ 10μM |
|
Z50592-2-O |
Oryctolagus Cuniculus (cluster #2 Of 8), Other |
Other |
9670 |
0.25 |
Functional ≤ 10μM |
|
Z80471-1-O |
SH-SY5 (Bone Marrow Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
77 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.07 |
-9.72 |
-436.08 |
5 |
18 |
-5 |
322 |
495.032 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.07 |
-7.43 |
-792.66 |
3 |
18 |
-7 |
327 |
493.016 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
1978 |
0.28 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
340 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.68 |
5.78 |
-10.77 |
1 |
6 |
0 |
73 |
418.297 |
5 |
↓
|
|
|
|
Analogs
-
25373069
-
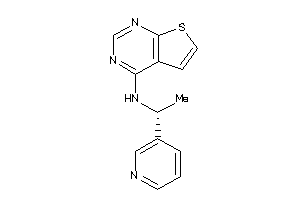
-
25373073
-
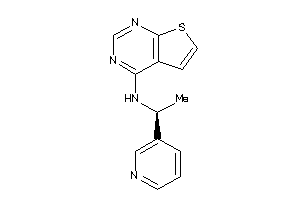
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
370 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
8.51 |
-7.57 |
1 |
3 |
0 |
38 |
255.346 |
3 |
↓
|
|
|
|
Analogs
-
31948961
-
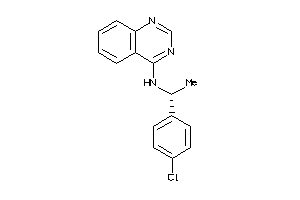
-
31948963
-
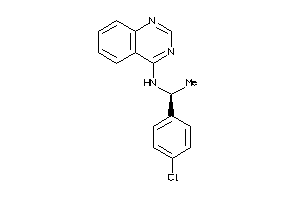
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
127 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
9.39 |
-7.9 |
1 |
3 |
0 |
38 |
249.317 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.48 |
9.85 |
-28.77 |
2 |
3 |
1 |
39 |
250.325 |
3 |
↓
|
|
|
|
Analogs
-
31948961
-

-
31948963
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
46 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
10.06 |
-7.87 |
1 |
3 |
0 |
38 |
263.344 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
41471380
-
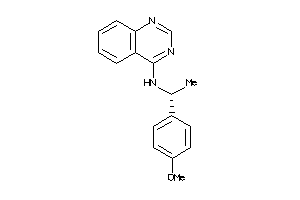
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
45 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
8.68 |
-9.35 |
1 |
4 |
0 |
47 |
279.343 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
44126678
-
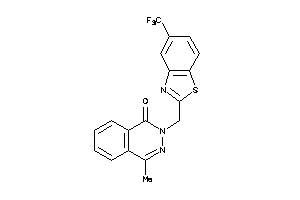
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
0.96 |
-59.14 |
0 |
6 |
-1 |
87 |
418.376 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
147 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
10.17 |
-7.35 |
1 |
3 |
0 |
38 |
263.344 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
500 |
0.37 |
Functional ≤ 10μM
|
|
Z80017-1-O |
A2780cisR (Cisplatin-resistant Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
60 |
0.42 |
Functional ≤ 10μM
|
|
Z80055-1-O |
CAKI-1 (Kidney Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
40 |
0.43 |
Functional ≤ 10μM
|
|
Z80089-1-O |
CHO-AA8 (cluster #1 Of 2), Other |
Other |
1 |
0.53 |
Functional ≤ 10μM
|
|
Z80114-1-O |
Daudi (Burkitts Lymphoma Cells) (cluster #1 Of 4), Other |
Other |
450 |
0.37 |
Functional ≤ 10μM
|
|
Z80115-1-O |
DC3F (cluster #1 Of 1), Other |
Other |
82 |
0.41 |
Functional ≤ 10μM
|
|
Z80117-1-O |
DC3F/AD-II (cluster #1 Of 2), Other |
Other |
110 |
0.41 |
Functional ≤ 10μM
|
|
Z80125-8-O |
DU-145 (Prostate Carcinoma) (cluster #8 Of 9), Other |
Other |
170 |
0.39 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
50 |
0.43 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
99 |
0.41 |
Functional ≤ 10μM |
|
Z80186-4-O |
K562 (Erythroleukemia Cells) (cluster #4 Of 11), Other |
Other |
470 |
0.37 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
90 |
0.41 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
960 |
0.35 |
Functional ≤ 10μM
|
|
Z80244-3-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #3 Of 7), Other |
Other |
270 |
0.38 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
51 |
0.43 |
Functional ≤ 10μM
|
|
Z80364-1-O |
P388/ADR (Lymphoma Cells) (cluster #1 Of 1), Other |
Other |
535 |
0.37 |
Functional ≤ 10μM
|
|
Z80367-1-O |
P388/S (cluster #1 Of 1), Other |
Other |
48 |
0.43 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
1500 |
0.34 |
Functional ≤ 10μM
|
|
Z80433-1-O |
RPMI-8226 (Multiple Myeloma Cells) (cluster #1 Of 3), Other |
Other |
200 |
0.39 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
993 |
0.35 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
200 |
0.39 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
7 |
0.48 |
Functional ≤ 10μM
|
|
Z80647-1-O |
833K Cell Line (cluster #1 Of 1), Other |
Other |
328 |
0.38 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
4500 |
0.31 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
400 |
0.37 |
Functional ≤ 10μM
|
|
Z80768-1-O |
CH1 (Ovarian Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
40 |
0.43 |
Functional ≤ 10μM
|
|
Z80776-1-O |
Clone 62 Cell Line (cluster #1 Of 1), Other |
Other |
1500 |
0.34 |
Functional ≤ 10μM
|
|
Z80888-1-O |
H2987 Cell Line (cluster #1 Of 1), Other |
Other |
1600 |
0.34 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
1700 |
0.34 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
10 |
0.47 |
Functional ≤ 10μM
|
|
Z81020-2-O |
HepG2 (Hepatoblastoma Cells) (cluster #2 Of 8), Other |
Other |
5400 |
0.31 |
Functional ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
50 |
0.43 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
600 |
0.36 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
810 |
0.36 |
Functional ≤ 10μM
|
|
Z81264-1-O |
V79 (Lung Fibroblasts) (cluster #1 Of 2), Other |
Other |
800 |
0.36 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
70 |
0.42 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 2), Other |
Other |
4030 |
0.31 |
ADME/T ≤ 10μM
|
|
Z81264-1-O |
V79 (Lung Fibroblasts) (cluster #1 Of 2), Other |
Other |
400 |
0.37 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.23 |
-4.19 |
-10.5 |
4 |
9 |
0 |
145 |
334.332 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.23 |
-4.19 |
-11.87 |
4 |
9 |
0 |
145 |
334.332 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.52 |
-3.22 |
-9.52 |
2 |
4 |
0 |
58 |
299.399 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
-3 |
-9.13 |
2 |
4 |
0 |
58 |
313.426 |
4 |
↓
|
|
|
|
Analogs
-
5430822
-
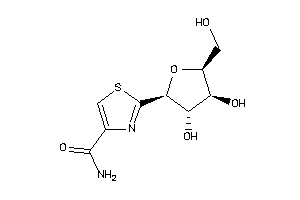
-
5430826
-
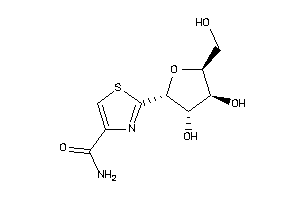
-
6091415
-
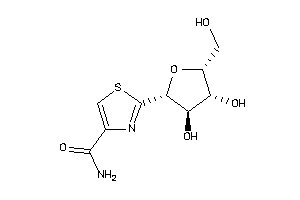
-
16941331
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
94 |
0.58 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
10000 |
0.41 |
Functional ≤ 10μM
|
|
Z80068-1-O |
CCRF-SB (Lymphoblastic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
7500 |
0.42 |
Functional ≤ 10μM
|
|
Z80092-1-O |
CHO-K1 (Ovarian Cells) (cluster #1 Of 2), Other |
Other |
6400 |
0.43 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
9320 |
0.41 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
1010 |
0.49 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
5290 |
0.43 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7400 |
0.42 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
8490 |
0.42 |
Functional ≤ 10μM
|
|
Z80285-1-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
5200 |
0.44 |
Functional ≤ 10μM
|
|
Z80291-1-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
850 |
0.50 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
2200 |
0.47 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
8200 |
0.42 |
Functional ≤ 10μM
|
|
Z80595-1-O |
WIL2-NS (Lymphoblastoid Cells) (cluster #1 Of 2), Other |
Other |
5700 |
0.43 |
Functional ≤ 10μM
|
|
Z81166-1-O |
LLC Cell Line (cluster #1 Of 1), Other |
Other |
1500 |
0.48 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
4760 |
0.44 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 4), Other |
Other |
1840 |
0.47 |
ADME/T ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
890 |
0.50 |
ADME/T ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 3), Other |
Other |
5290 |
0.43 |
ADME/T ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
6390 |
0.43 |
ADME/T ≤ 10μM
|
|
Z80291-1-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
850 |
0.50 |
ADME/T ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 2), Other |
Other |
510 |
0.52 |
ADME/T ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
4760 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.52 |
-8.8 |
-12.79 |
5 |
7 |
0 |
126 |
260.271 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTH1R-1-E |
Parathyroid Hormone Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.38 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
1500 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.58 |
4.7 |
-8.46 |
4 |
7 |
0 |
113 |
282.344 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
0.71 |
-10.43 |
0 |
3 |
0 |
33 |
352.865 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
2.07 |
-53.98 |
1 |
4 |
0 |
53 |
327.424 |
10 |
↓
|
|
|
|
|
|
|
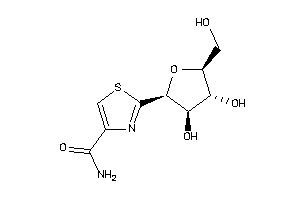
Analogs
-
35824094
-
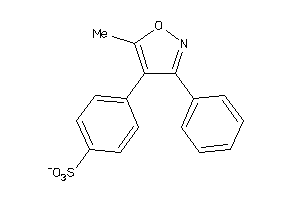
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
180 |
0.41 |
Binding ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
329 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
1.09 |
-15.89 |
3 |
6 |
0 |
106 |
330.365 |
4 |
↓
|
|
|
|
Analogs
-
13524645
-
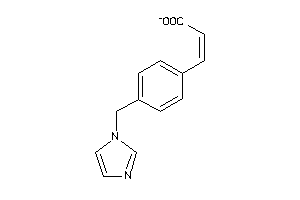
Draw
Identity
99%
90%
80%
70%
Vendors
And 33 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
9.62 |
-54.16 |
0 |
4 |
-1 |
58 |
227.243 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.56 |
10.13 |
-73.47 |
1 |
4 |
0 |
59 |
228.251 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
140 |
0.37 |
Binding ≤ 10μM
|
|
CYNT-1-B |
Carbonic Anhydrase (cluster #1 Of 3), Bacterial |
Bacteria |
713 |
0.33 |
Binding ≤ 10μM
|
|
B5SU02-2-E |
Alpha Carbonic Anhydrase (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
34 |
0.40 |
Binding ≤ 10μM
|
|
C0IX24-1-E |
Carbonic Anhydrase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
690 |
0.33 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
18 |
0.42 |
Binding ≤ 10μM |
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
98 |
0.38 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
689 |
0.33 |
Binding ≤ 10μM
|
|
CAH15-2-E |
Carbonic Anhydrase 15 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
45 |
0.40 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
21 |
0.41 |
Binding ≤ 10μM |
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
290 |
0.35 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
794 |
0.33 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
93 |
0.38 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
94 |
0.38 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2170 |
0.30 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
16 |
0.42 |
Binding ≤ 10μM |
|
COX2-1-E |
Cytochrome C Oxidase Subunit 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.39 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
810 |
0.33 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9730 |
0.27 |
Binding ≤ 10μM
|
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9 |
0.43 |
Binding ≤ 10μM
|
|
Q8HZR1-1-E |
Cyclooxygenase-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5570 |
0.28 |
Binding ≤ 10μM
|
|
Q8SPQ9-2-E |
Cyclooxygenase-2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.33 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
21 |
0.41 |
Functional ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.35 |
Functional ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.29 |
Functional ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
ADME/T ≤ 10μM
|
|
CAN-1-F |
Carbonic Anhydrase (cluster #1 Of 3), Fungal |
Fungi |
108 |
0.38 |
Binding ≤ 10μM
|
|
Q5AJ71-1-F |
Carbonic Anhydrase (cluster #1 Of 4), Fungal |
Fungi |
21 |
0.41 |
Binding ≤ 10μM
|
|
Z100741-1-O |
MC9 (Mast Cells) (cluster #1 Of 2), Other |
Other |
400 |
0.34 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
6670 |
0.28 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
5000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
5.33 |
-11.94 |
2 |
5 |
0 |
78 |
381.379 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
-3.5 |
-72.59 |
1 |
6 |
0 |
69 |
355.438 |
7 |
↓
|
|
|
|
Analogs
-
34956345
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
5.54 |
-8.66 |
0 |
3 |
0 |
39 |
239.303 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
9000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
-1.28 |
-10.56 |
1 |
4 |
0 |
51 |
336.416 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
7 |
0.42 |
Binding ≤ 10μM
|
|
O30463-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
1500 |
0.30 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.42 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
2 |
0.45 |
Binding ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
27 |
0.39 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
45 |
0.38 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
20 |
0.40 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
770 |
0.32 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
2 |
0.45 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
3 |
0.44 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
7 |
0.42 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
8 |
0.42 |
Functional ≤ 10μM
|
|
Z80459-1-O |
SCC-7 (cluster #1 Of 1), Other |
Other |
6 |
0.43 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
11 |
0.41 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
16 |
0.40 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
5 |
0.43 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
60 |
0.37 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
58 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
5.85 |
-37.21 |
6 |
8 |
1 |
119 |
370.433 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.19 |
5.38 |
-15.09 |
5 |
8 |
0 |
118 |
369.425 |
6 |
↓
|
|
|
|
Analogs
-
25457273
-
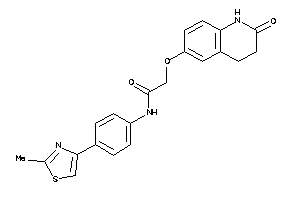
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
600 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.15 |
-0.94 |
-12.44 |
1 |
5 |
0 |
60 |
380.469 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
7.34 |
-8.34 |
0 |
2 |
0 |
26 |
238.315 |
2 |
↓
|
|
|
|
Analogs
-
3783756
-
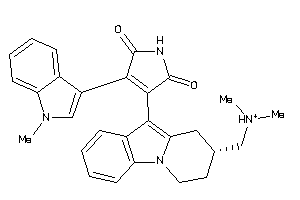
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
20 |
0.32 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.22 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5900 |
0.22 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5900 |
0.22 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
22 |
0.32 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
31 |
0.31 |
Binding ≤ 10μM |
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
42 |
0.30 |
Binding ≤ 10μM
|
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM |
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCD1-1-E |
Protein Kinase C Mu (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM |
|
KPCD1-1-E |
Protein Kinase C Mu (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCD3-1-E |
Protein Kinase C Nu (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM |
|
KPCD3-1-E |
Protein Kinase C Nu (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM |
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
37 |
0.31 |
Binding ≤ 10μM |
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
42 |
0.30 |
Binding ≤ 10μM
|
|
KPCI-1-E |
Protein Kinase C Iota (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM |
|
KPCI-1-E |
Protein Kinase C Iota (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
21 |
0.32 |
Binding ≤ 10μM |
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
42 |
0.30 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
21 |
0.32 |
Binding ≤ 10μM |
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
42 |
0.30 |
Binding ≤ 10μM
|
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM |
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
32 |
0.31 |
Binding ≤ 10μM
|
|
PHKG1-1-E |
Phosphorylase Kinase Gamma Subunit 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Binding ≤ 10μM
|
|
PHKG2-1-E |
Phosphorylase Kinase Gamma Subunit 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCD1-1-E |
Protein Kinase C Mu (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCD3-1-E |
Protein Kinase C Nu (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCL-1-E |
Protein Kinase C Eta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
950 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
13.61 |
-50.72 |
2 |
6 |
1 |
64 |
453.566 |
4 |
↓
|
|
|
|
Analogs
-
591953
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.22 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5900 |
0.22 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5900 |
0.22 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
42 |
0.30 |
Binding ≤ 10μM
|
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCD1-1-E |
Protein Kinase C Mu (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCD3-1-E |
Protein Kinase C Nu (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
42 |
0.30 |
Binding ≤ 10μM
|
|
KPCI-1-E |
Protein Kinase C Iota (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.30 |
Binding ≤ 10μM
|
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
42 |
0.30 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
42 |
0.30 |
Binding ≤ 10μM
|
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
32 |
0.31 |
Binding ≤ 10μM
|
|
PHKG1-1-E |
Phosphorylase Kinase Gamma Subunit 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Binding ≤ 10μM
|
|
PHKG2-1-E |
Phosphorylase Kinase Gamma Subunit 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCD1-1-E |
Protein Kinase C Mu (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCD3-1-E |
Protein Kinase C Nu (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCL-1-E |
Protein Kinase C Eta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
950 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
13.56 |
-48.59 |
2 |
6 |
1 |
64 |
453.566 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
300 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
7.51 |
-13.19 |
1 |
5 |
0 |
60 |
366.442 |
4 |
↓
|
|
|
|
Analogs
-
4073970
-
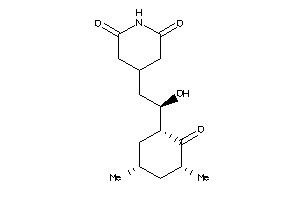
-
4536598
-
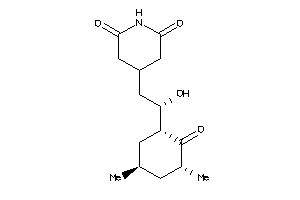
-
8579261
-
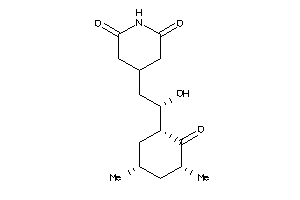
-
11592912
-
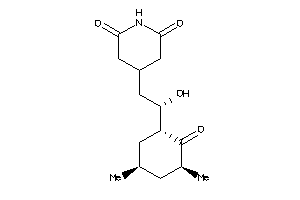
-
11592913
-
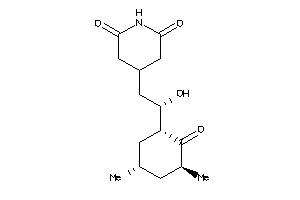
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.38 |
Binding ≤ 10μM
|
|
Z50347-2-O |
Saccharomyces Cerevisiae (cluster #2 Of 3), Other |
Other |
88 |
0.49 |
Functional ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
200 |
0.47 |
Functional ≤ 10μM
|
|
Z50466-5-O |
Trypanosoma Cruzi (cluster #5 Of 8), Other |
Other |
400 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
930 |
0.42 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
5000 |
0.37 |
Functional ≤ 10μM
|
|
Z80026-2-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
3600 |
0.38 |
Functional ≤ 10μM |
|
Z80224-6-O |
MCF7 (Breast Carcinoma Cells) (cluster #6 Of 14), Other |
Other |
1500 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
3500 |
0.38 |
Functional ≤ 10μM
|
|
Z80485-2-O |
SK-MEL-28 (Melanoma Cells) (cluster #2 Of 6), Other |
Other |
1000 |
0.42 |
Functional ≤ 10μM
|
|
Z80490-1-O |
SK-MES-1 (cluster #1 Of 4), Other |
Other |
2700 |
0.39 |
Functional ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 7), Other |
Other |
530 |
0.44 |
Functional ≤ 10μM
|
|
Z80653-1-O |
9L (Glioma Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.47 |
Functional ≤ 10μM
|
|
Z80874-4-O |
CEM (T-cell Leukemia) (cluster #4 Of 7), Other |
Other |
80 |
0.50 |
Functional ≤ 10μM
|
|
Z81020-4-O |
HepG2 (Hepatoblastoma Cells) (cluster #4 Of 8), Other |
Other |
2500 |
0.39 |
Functional ≤ 10μM
|
|
Z81072-7-O |
Jurkat (Acute Leukemic T-cells) (cluster #7 Of 10), Other |
Other |
930 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-3-O |
LNCaP (Prostate Carcinoma) (cluster #3 Of 5), Other |
Other |
1200 |
0.41 |
Functional ≤ 10μM
|
|
Z81247-6-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #6 Of 9), Other |
Other |
533 |
0.44 |
Functional ≤ 10μM
|
|
Z81252-4-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #4 Of 11), Other |
Other |
1200 |
0.41 |
Functional ≤ 10μM |
|
Z80005-2-O |
3T3 (Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
912 |
0.42 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
3.22 |
-17 |
2 |
5 |
0 |
83 |
281.352 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
5 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
10.03 |
-55.46 |
0 |
7 |
-1 |
101 |
437.431 |
7 |
↓
|
|