|
|
Analogs
-
33955165
-
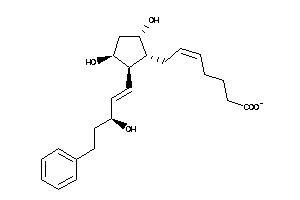
-
33955166
-
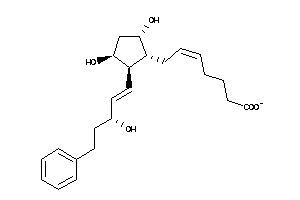
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50589-1-O |
Felis Catus (cluster #1 Of 2), Other |
Other |
1 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
6.16 |
-52.89 |
3 |
5 |
-1 |
101 |
387.496 |
11 |
↓
|
|
|
|
Analogs
-
33955165
-

-
33955166
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50589-1-O |
Felis Catus (cluster #1 Of 2), Other |
Other |
1 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
5.94 |
-53.13 |
3 |
5 |
-1 |
101 |
387.496 |
11 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.66 |
6.07 |
-13.76 |
1 |
5 |
0 |
59 |
240.266 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.66 |
6.59 |
-44.63 |
2 |
5 |
1 |
61 |
241.274 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.41 |
3.6 |
-18.77 |
2 |
6 |
0 |
94 |
241.254 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.05 |
1.58 |
-44.81 |
1 |
6 |
-1 |
98 |
240.246 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.41 |
4.07 |
-49.53 |
3 |
6 |
1 |
96 |
242.262 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
E1BN64-1-E |
Phosphodiesterase 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.53 |
Binding ≤ 10μM
|
|
O77823-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.46 |
Binding ≤ 10μM
|
|
PDE3A-2-E |
Phosphodiesterase 3A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.53 |
Binding ≤ 10μM |
|
PDE3B-2-E |
Phosphodiesterase 3B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.53 |
Binding ≤ 10μM |
|
PDE4A-3-E |
Phosphodiesterase 4A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE4B-2-E |
Phosphodiesterase 4B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE4C-2-E |
Phosphodiesterase 4C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE4D-2-E |
Phosphodiesterase 4D (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE5A-2-E |
Phosphodiesterase 5A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.46 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.54 |
Binding ≤ 10μM
|
|
Q9XSW7-1-E |
Phosphodiesterase 3A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
843 |
0.53 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z102213-1-O |
Blood (cluster #1 Of 2), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
122 |
0.60 |
Functional ≤ 10μM
|
|
Z50512-5-O |
Cavia Porcellus (cluster #5 Of 7), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
9000 |
0.44 |
Functional ≤ 10μM
|
|
Z50588-6-O |
Canis Familiaris (cluster #6 Of 7), Other |
Other |
700 |
0.54 |
Functional ≤ 10μM
|
|
Z50589-1-O |
Felis Catus (cluster #1 Of 2), Other |
Other |
7700 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
4.19 |
-16.01 |
1 |
4 |
0 |
70 |
211.224 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.40 |
2.16 |
-43.19 |
0 |
4 |
-1 |
73 |
210.216 |
1 |
↓
|
|
|
|
Analogs
-
4095842
-
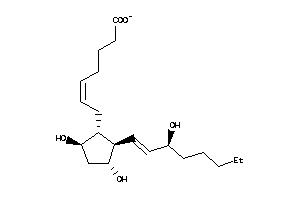
-
8552290
-
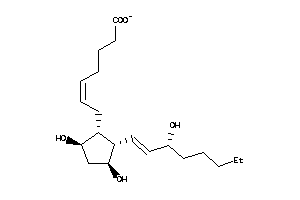
-
8552291
-
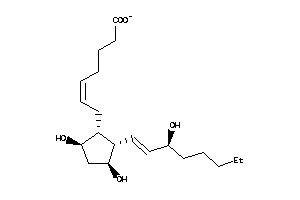
-
8552292
-
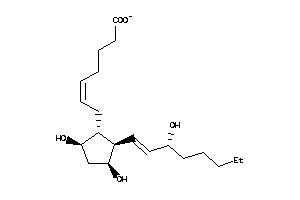
-
8552293
-
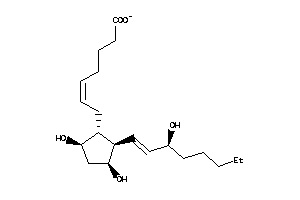
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PD2R-2-E |
Prostanoid DP Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.30 |
Binding ≤ 10μM
|
|
PE2R1-2-E |
Prostanoid EP1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
380 |
0.36 |
Binding ≤ 10μM
|
|
PE2R3-2-E |
Prostanoid EP3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
322 |
0.36 |
Binding ≤ 10μM
|
|
PE2R4-2-E |
Prostanoid EP4 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.30 |
Binding ≤ 10μM
|
|
PF2R-1-E |
Prostaglandin F2-alpha Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
PE2R1-2-E |
Prostanoid EP1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.35 |
Functional ≤ 10μM
|
|
PE2R3-2-E |
Prostanoid EP3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.32 |
Functional ≤ 10μM
|
|
PF2R-1-E |
Prostaglandin F2-alpha Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.43 |
Functional ≤ 10μM
|
|
TA2R-1-E |
Thromboxane A2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.33 |
Functional ≤ 10μM
|
|
Z50589-1-O |
Felis Catus (cluster #1 Of 2), Other |
Other |
7 |
0.46 |
Functional ≤ 10μM
|
|
Z80530-2-O |
Swiss-3T3 (Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
25 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
5.6 |
-49.94 |
3 |
5 |
-1 |
101 |
353.479 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.55 |
-2.55 |
-8.97 |
2 |
4 |
0 |
67 |
132.115 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.05 |
3.63 |
-9.76 |
4 |
5 |
0 |
98 |
354.487 |
12 |
↓
|
|