|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101843-2-O |
Human Coxsackievirus B2 (cluster #2 Of 3), Other |
Other |
3790 |
0.28 |
Functional ≤ 10μM
|
|
Z101846-1-O |
Human Echovirus 6 (cluster #1 Of 2), Other |
Other |
9 |
0.42 |
Functional ≤ 10μM
|
|
Z101847-1-O |
Human Enterovirus 68 (cluster #1 Of 1), Other |
Other |
3470 |
0.28 |
Functional ≤ 10μM
|
|
Z103013-1-O |
Human Coxsackievirus A9 (cluster #1 Of 2), Other |
Other |
3790 |
0.28 |
Functional ≤ 10μM
|
|
Z50615-1-O |
Human Echovirus 9 (cluster #1 Of 2), Other |
Other |
9 |
0.42 |
Functional ≤ 10μM
|
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
3790 |
0.28 |
Functional ≤ 10μM
|
|
Z50618-1-O |
Human Coxsackievirus B5 (cluster #1 Of 2), Other |
Other |
3790 |
0.28 |
Functional ≤ 10μM
|
|
Z50667-1-O |
Human Coxsackievirus B1 (cluster #1 Of 2), Other |
Other |
3790 |
0.28 |
Functional ≤ 10μM
|
|
Z50668-1-O |
Human Coxsackievirus B4 (cluster #1 Of 2), Other |
Other |
3790 |
0.28 |
Functional ≤ 10μM
|
|
Z50669-1-O |
Human Echovirus 11 (cluster #1 Of 1), Other |
Other |
9 |
0.42 |
Functional ≤ 10μM
|
|
Z50731-1-O |
Echovirus (cluster #1 Of 1), Other |
Other |
9 |
0.42 |
Functional ≤ 10μM
|
|
Z50746-1-O |
Human Enterovirus 71 (cluster #1 Of 2), Other |
Other |
3470 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.16 |
1.38 |
-8.98 |
0 |
3 |
0 |
27 |
423.723 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
4900 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
3.5 |
-10.08 |
0 |
5 |
0 |
78 |
309.108 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
36985165
-
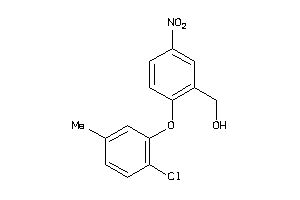
-
36985167
-
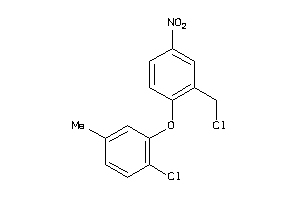
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
6900 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
3.63 |
-9.32 |
0 |
5 |
0 |
78 |
274.663 |
3 |
↓
|
|
|
|
Analogs
-
3506330
-
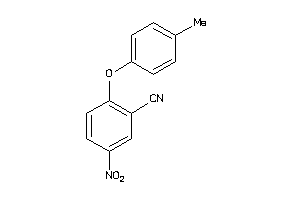
-
8063814
-
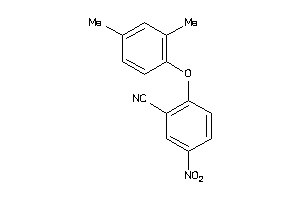
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
4100 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
5.13 |
-12.07 |
0 |
6 |
0 |
102 |
265.228 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50078-1-O |
Human Rhinovirus 45 (cluster #1 Of 1), Other |
Other |
250 |
0.37 |
Functional ≤ 10μM
|
|
Z50085-1-O |
Human Rhinovirus Sp. (cluster #1 Of 1), Other |
Other |
40 |
0.41 |
Functional ≤ 10μM
|
|
Z50088-1-O |
Human Rhinovirus 9 (cluster #1 Of 1), Other |
Other |
38 |
0.42 |
Functional ≤ 10μM
|
|
Z50091-1-O |
Human Rhinovirus 41 (cluster #1 Of 1), Other |
Other |
27 |
0.42 |
Functional ≤ 10μM
|
|
Z50093-1-O |
Human Rhinovirus 63 (cluster #1 Of 1), Other |
Other |
27 |
0.42 |
Functional ≤ 10μM
|
|
Z50094-1-O |
Human Rhinovirus 70 (cluster #1 Of 1), Other |
Other |
110 |
0.39 |
Functional ≤ 10μM
|
|
Z50095-1-O |
Human Rhinovirus 86 (cluster #1 Of 1), Other |
Other |
140 |
0.38 |
Functional ≤ 10μM
|
|
Z50130-1-O |
Human Rhinovirus 15 (cluster #1 Of 1), Other |
Other |
42 |
0.41 |
Functional ≤ 10μM
|
|
Z50132-1-O |
Human Rhinovirus 39 (cluster #1 Of 1), Other |
Other |
28 |
0.42 |
Functional ≤ 10μM
|
|
Z50135-1-O |
Human Rhinovirus 85 (cluster #1 Of 1), Other |
Other |
33 |
0.42 |
Functional ≤ 10μM
|
|
Z50617-1-O |
Human Coxsackievirus B3 (cluster #1 Of 2), Other |
Other |
700 |
0.34 |
Functional ≤ 10μM
|
|
Z50620-1-O |
Human Poliovirus 3 (cluster #1 Of 1), Other |
Other |
40 |
0.41 |
Functional ≤ 10μM
|
|
Z50622-1-O |
Human Rhinovirus 1B (cluster #1 Of 2), Other |
Other |
40 |
0.41 |
Functional ≤ 10μM
|
|
Z50670-1-O |
Human Poliovirus 2 (cluster #1 Of 1), Other |
Other |
60 |
0.40 |
Functional ≤ 10μM
|
|
Z50674-1-O |
Human Rhinovirus 2 (cluster #1 Of 1), Other |
Other |
26 |
0.42 |
Functional ≤ 10μM
|
|
Z50675-1-O |
Human Rhinovirus 89 (cluster #1 Of 1), Other |
Other |
100 |
0.39 |
Functional ≤ 10μM
|
|
Z50679-2-O |
Human Rhinovirus 14 (cluster #2 Of 2), Other |
Other |
110 |
0.39 |
Functional ≤ 10μM
|
|
Z50792-1-O |
Human Rhinovirus 29 (cluster #1 Of 1), Other |
Other |
29 |
0.42 |
Functional ≤ 10μM
|
|
Z50793-1-O |
Human Rhinovirus 72 (cluster #1 Of 1), Other |
Other |
720 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
5.22 |
-13.89 |
3 |
7 |
0 |
111 |
358.423 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.57 |
5.22 |
-14.35 |
3 |
7 |
0 |
111 |
358.423 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
5000 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
3.59 |
-8.71 |
0 |
5 |
0 |
78 |
274.663 |
3 |
↓
|
|
|
|
Analogs
-
8063814
-

-
2623253
-
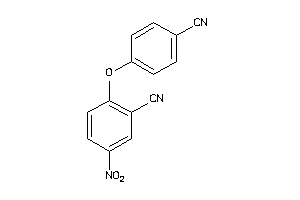
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
6200 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
4.06 |
-8.6 |
0 |
5 |
0 |
78 |
254.245 |
3 |
↓
|
|
|
|
Analogs
-
34978321
-
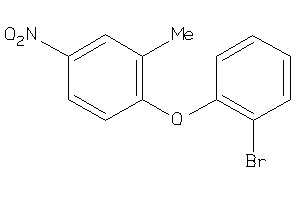
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
5700 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
2.98 |
-8.7 |
0 |
5 |
0 |
78 |
319.114 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.62 |
3.46 |
-8.43 |
0 |
5 |
0 |
78 |
309.108 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
4900 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
3.59 |
-9.13 |
0 |
5 |
0 |
78 |
274.663 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-1-O |
Human Coxsackievirus B3 (cluster #1 Of 2), Other |
Other |
459 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
7.29 |
-22.76 |
2 |
5 |
0 |
71 |
314.348 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
3800 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.62 |
3.48 |
-8.31 |
0 |
5 |
0 |
78 |
309.108 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-1-O |
Human Coxsackievirus B3 (cluster #1 Of 2), Other |
Other |
1630 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
7.41 |
-19.95 |
2 |
5 |
0 |
71 |
332.338 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
2000 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.39 |
3.91 |
-7.89 |
0 |
5 |
0 |
78 |
288.69 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
5000 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
2.98 |
-9.12 |
0 |
5 |
0 |
78 |
319.114 |
3 |
↓
|
|
|
|
Analogs
-
37987972
-
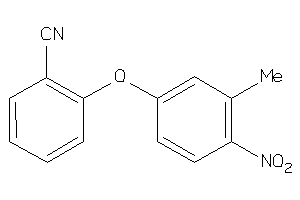
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
7400 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
5.12 |
-12.38 |
0 |
6 |
0 |
102 |
265.228 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
3700 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.75 |
2.87 |
-8.31 |
0 |
5 |
0 |
78 |
353.559 |
3 |
↓
|
|
|
|
Analogs
-
34978321
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
6200 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
2.28 |
-8.22 |
0 |
5 |
0 |
78 |
398.01 |
3 |
↓
|
|
|
|
Analogs
-
8148827
-
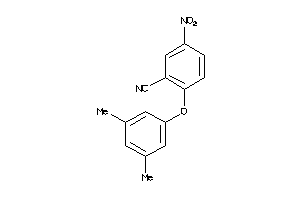
-
37987972
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
6400 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.76 |
4.07 |
-7.97 |
0 |
5 |
0 |
78 |
254.245 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
2600 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.52 |
9.64 |
-7.76 |
0 |
5 |
0 |
79 |
333.141 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
5700 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
2.91 |
-8.47 |
0 |
5 |
0 |
78 |
366.114 |
3 |
↓
|
|
|
|
Analogs
-
34978321
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
3700 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.75 |
9.57 |
-8.35 |
0 |
5 |
0 |
79 |
353.559 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50617-2-O |
Human Coxsackievirus B3 (cluster #2 Of 2), Other |
Other |
5700 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.39 |
2.91 |
-8.96 |
0 |
5 |
0 |
78 |
366.114 |
3 |
↓
|
|