|
|
Analogs
-
4212843
-
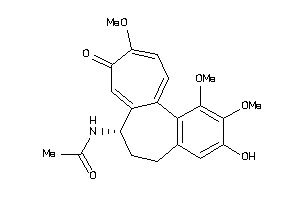
-
607790
-
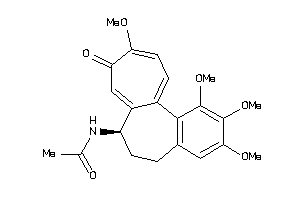
Draw
Identity
99%
90%
80%
70%
Vendors
And 53 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q862F3-2-E |
Tubulin Alpha Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.27 |
Binding ≤ 10μM |
|
Q862L2-2-E |
Tubulin Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.27 |
Binding ≤ 10μM |
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.27 |
Binding ≤ 10μM
|
|
TBB-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1260 |
0.28 |
Binding ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.29 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.29 |
Binding ≤ 10μM
|
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.30 |
Binding ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.30 |
Binding ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.30 |
Binding ≤ 10μM
|
|
TBG1-1-E |
Tubulin Gamma-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.24 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.24 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.29 |
Functional ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.28 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.29 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.24 |
Functional ≤ 10μM
|
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.26 |
Functional ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
22 |
0.37 |
Functional ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
20 |
0.37 |
Functional ≤ 10μM |
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
53 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
1400 |
0.28 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
18 |
0.37 |
Functional ≤ 10μM |
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
12 |
0.38 |
Functional ≤ 10μM
|
|
Z80024-1-O |
A9 (Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.36 |
Functional ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 7), Other |
Other |
31 |
0.36 |
Functional ≤ 10μM
|
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
220 |
0.32 |
Functional ≤ 10μM |
|
Z80096-1-O |
CHO-TAX 5-6 (cluster #1 Of 2), Other |
Other |
140 |
0.33 |
Functional ≤ 10μM |
|
Z80097-1-O |
CHO-VV 3-2 (cluster #1 Of 2), Other |
Other |
620 |
0.30 |
Functional ≤ 10μM |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
190 |
0.32 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
10 |
0.39 |
Functional ≤ 10μM |
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
53 |
0.35 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
1 |
0.43 |
Functional ≤ 10μM |
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
22 |
0.37 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1900 |
0.28 |
Functional ≤ 10μM |
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
15 |
0.38 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
800 |
0.29 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
10 |
0.39 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
33 |
0.36 |
Functional ≤ 10μM |
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM
|
|
Z80427-1-O |
RKO (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
20 |
0.37 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM |
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
5 |
0.40 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
71 |
0.35 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
30 |
0.36 |
Functional ≤ 10μM |
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
5 |
0.40 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
390 |
0.31 |
Functional ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
45 |
0.35 |
Functional ≤ 10μM
|
|
Z80711-1-O |
NCI/ADR-RES (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
1113 |
0.29 |
Functional ≤ 10μM |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
9 |
0.39 |
Functional ≤ 10μM
|
|
Z80720-1-O |
Burkitts Lymphoma Cells (cluster #1 Of 1), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
52 |
0.35 |
Functional ≤ 10μM |
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
3 |
0.41 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
8 |
0.39 |
Functional ≤ 10μM |
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
40 |
0.36 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM |
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
70 |
0.35 |
Functional ≤ 10μM |
|
Z81054-2-O |
SF-268 (Glioblastoma Cells) (cluster #2 Of 3), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
17 |
0.38 |
Functional ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
20 |
0.37 |
Functional ≤ 10μM |
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
93 |
0.34 |
Functional ≤ 10μM |
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
39 |
0.36 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
70 |
0.35 |
Functional ≤ 10μM
|
|
Z81281-1-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 5), Other |
Other |
13 |
0.38 |
Functional ≤ 10μM |
|
Z81283-1-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM |
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM |
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
140 |
0.33 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 3), Other |
Other |
530 |
0.30 |
ADME/T ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
522 |
0.30 |
ADME/T ≤ 10μM
|
|
Z80897-1-O |
H9 (T-lymphoid Cells) (cluster #1 Of 2), Other |
Other |
10 |
0.39 |
ADME/T ≤ 10μM |
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.41 |
ADME/T ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 3), Other |
Other |
210 |
0.32 |
ADME/T ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
2 |
0.42 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
7.99 |
-20.74 |
1 |
7 |
0 |
83 |
399.443 |
5 |
↓
|
|
|
|
Analogs
-
3978074
-
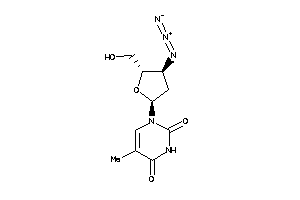
-
4245659
-
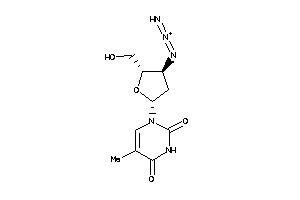
-
4245660
-
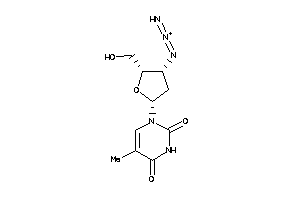
-
4467870
-
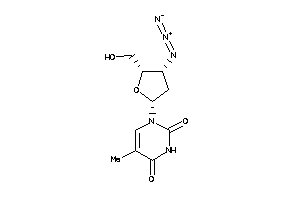
-
11592709
-
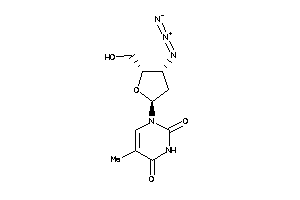
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KITH-1-E |
Thymidine Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Binding ≤ 10μM
|
|
CXCR4-1-E |
C-X-C Chemokine Receptor Type 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.60 |
Functional ≤ 10μM
|
|
Z80295-1-O |
MT4 (Lymphocytes) (cluster #1 Of 1), Other |
Other |
20 |
0.57 |
Binding ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
4 |
0.62 |
Functional ≤ 10μM
|
|
Z102020-1-O |
Porcine Endogenous Retrovirus (cluster #1 Of 1), Other |
Other |
23 |
0.56 |
Functional ≤ 10μM
|
|
Z50459-2-O |
Leishmania Donovani (cluster #2 Of 8), Other |
Other |
3400 |
0.40 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
7 |
0.60 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
46 |
0.54 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
9600 |
0.37 |
Functional ≤ 10μM
|
|
Z50609-1-O |
Simian Immunodeficiency Virus (cluster #1 Of 2), Other |
Other |
12 |
0.58 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
9 |
0.59 |
Functional ≤ 10μM
|
|
Z50664-1-O |
Rauscher Murine Leukemia Virus (cluster #1 Of 1), Other |
Other |
23 |
0.56 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
9 |
0.59 |
Functional ≤ 10μM
|
|
Z50737-1-O |
Human Immunodeficiency Virus 3 (cluster #1 Of 1), Other |
Other |
40 |
0.55 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
9 |
0.59 |
Functional ≤ 10μM
|
|
Z80071-1-O |
CEM-0 (T-cell Leukemia) (cluster #1 Of 2), Other |
Other |
6 |
0.61 |
Functional ≤ 10μM
|
|
Z80073-1-O |
CEM-c113 (cluster #1 Of 1), Other |
Other |
6 |
0.61 |
Functional ≤ 10μM
|
|
Z80076-1-O |
CEM-SS (T-cell Leukemia) (cluster #1 Of 5), Other |
Other |
95 |
0.52 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
6180 |
0.38 |
Functional ≤ 10μM
|
|
Z80285-1-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
6 |
0.61 |
Functional ≤ 10μM
|
|
Z80294-1-O |
MT2 (Lymphocytes) (cluster #1 Of 1), Other |
Other |
6 |
0.61 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
10 |
0.59 |
Functional ≤ 10μM
|
|
Z80384-1-O |
PBL (Peripheral Blood Lymphocytes) (cluster #1 Of 1), Other |
Other |
29 |
0.56 |
Functional ≤ 10μM
|
|
Z80516-1-O |
SUP-T1 (cluster #1 Of 1), Other |
Other |
50 |
0.54 |
Functional ≤ 10μM |
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
40 |
0.55 |
Functional ≤ 10μM
|
|
Z80683-1-O |
ATH8 Cell Line (cluster #1 Of 1), Other |
Other |
400 |
0.47 |
Functional ≤ 10μM
|
|
Z80688-1-O |
B(EBV+) Cell Line (cluster #1 Of 1), Other |
Other |
1 |
0.66 |
Functional ≤ 10μM
|
|
Z80721-1-O |
BxT Cell Line (cluster #1 Of 1), Other |
Other |
55 |
0.53 |
Functional ≤ 10μM
|
|
Z80737-1-O |
C3H/3T3 (Embryonic Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
20 |
0.57 |
Functional ≤ 10μM
|
|
Z80760-1-O |
CFU-GM Cell Line (cluster #1 Of 1), Other |
Other |
1500 |
0.43 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
870 |
0.45 |
Functional ≤ 10μM
|
|
Z80897-1-O |
H9 (T-lymphoid Cells) (cluster #1 Of 2), Other |
Other |
9 |
0.59 |
Functional ≤ 10μM |
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
10 |
0.59 |
Functional ≤ 10μM
|
|
Z81223-1-O |
M Cell Line (cluster #1 Of 1), Other |
Other |
15 |
0.58 |
Functional ≤ 10μM
|
|
Z81247-7-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #7 Of 9), Other |
Other |
9 |
0.59 |
Functional ≤ 10μM
|
|
Z80874-3-O |
CEM (T-cell Leukemia) (cluster #3 Of 4), Other |
Other |
700 |
0.45 |
ADME/T ≤ 10μM
|
|
Z80897-1-O |
H9 (T-lymphoid Cells) (cluster #1 Of 2), Other |
Other |
45 |
0.54 |
ADME/T ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
6 |
0.61 |
Binding ≤ 10μM
|
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
6 |
0.61 |
Binding ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 1), Viral |
Viruses |
8 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.10 |
-3.76 |
-14.65 |
2 |
9 |
0 |
134 |
267.245 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80035-5-O |
B16 (Melanoma Cells) (cluster #5 Of 7), Other |
Other |
269 |
0.40 |
Functional ≤ 10μM
|
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
218 |
0.41 |
Functional ≤ 10μM
|
|
Z80193-1-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #1 Of 12), Other |
Other |
415 |
0.39 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
185 |
0.41 |
Functional ≤ 10μM
|
|
Z80224-12-O |
MCF7 (Breast Carcinoma Cells) (cluster #12 Of 14), Other |
Other |
19 |
0.47 |
Functional ≤ 10μM
|
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
47 |
0.45 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
338 |
0.39 |
Functional ≤ 10μM
|
|
Z80461-1-O |
SCL-1 (cluster #1 Of 1), Other |
Other |
383 |
0.39 |
Functional ≤ 10μM
|
|
Z80475-3-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #3 Of 3), Other |
Other |
31 |
0.46 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
320 |
0.40 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
213 |
0.41 |
Functional ≤ 10μM
|
|
Z80901-1-O |
HaCaT (Keratinocytes) (cluster #1 Of 2), Other |
Other |
47 |
0.45 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
192 |
0.41 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
470 |
0.39 |
Functional ≤ 10μM
|
|
Z81252-7-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #7 Of 11), Other |
Other |
428 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.13 |
1.81 |
-10.68 |
1 |
5 |
0 |
65 |
316.353 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
21 |
0.43 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
21 |
0.43 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
21 |
0.43 |
Binding ≤ 10μM
|
|
CCND3-1-E |
G1/S-specific Cyclin D3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
356 |
0.36 |
Binding ≤ 10μM
|
|
CCNH-1-E |
Cyclin H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
126 |
0.39 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
21 |
0.43 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
CDK6-1-E |
Cyclin-dependent Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
356 |
0.36 |
Binding ≤ 10μM
|
|
CDK7-1-E |
Cyclin-dependent Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
126 |
0.39 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
20 |
0.43 |
Binding ≤ 10μM
|
|
KC1A-1-E |
Casein Kinase I Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3050 |
0.31 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3050 |
0.31 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3050 |
0.31 |
Binding ≤ 10μM
|
|
KC1G1-1-E |
Casein Kinase I Gamma 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3050 |
0.31 |
Binding ≤ 10μM
|
|
KC1G2-1-E |
Casein Kinase I Gamma 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3050 |
0.31 |
Binding ≤ 10μM
|
|
VGFR2-2-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.31 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
3 |
0.48 |
Binding ≤ 10μM
|
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
308 |
0.36 |
Binding ≤ 10μM
|
|
Z80018-2-O |
A-375 (Malignant Melanoma Cells) (cluster #2 Of 4), Other |
Other |
28 |
0.42 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
31 |
0.42 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
23 |
0.43 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
33 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
-4.76 |
-36.48 |
4 |
7 |
1 |
97 |
357.825 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
8.11 |
-13.19 |
1 |
4 |
0 |
51 |
342.395 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
12 |
0.26 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.28 |
Binding ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
4 |
0.28 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
9000 |
0.17 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.26 |
16.74 |
-128.39 |
4 |
9 |
2 |
102 |
578.643 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.26 |
12.92 |
-18.11 |
2 |
9 |
0 |
99 |
576.627 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.26 |
15.32 |
-53.34 |
3 |
9 |
1 |
100 |
577.635 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.25 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.27 |
Binding ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
11 |
0.27 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
10000 |
0.17 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.26 |
16.64 |
-136.87 |
4 |
9 |
2 |
102 |
578.643 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
4.33 |
12.99 |
-49.42 |
3 |
9 |
1 |
104 |
577.635 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.26 |
12.91 |
-17.97 |
2 |
9 |
0 |
99 |
576.627 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.32 |
9.18 |
-41.96 |
3 |
9 |
1 |
125 |
482.52 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
5.32 |
9.03 |
-36.32 |
3 |
9 |
1 |
125 |
482.52 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.32 |
9.8 |
-16.61 |
2 |
9 |
0 |
127 |
481.512 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.83 |
4.16 |
-15.65 |
0 |
5 |
0 |
65 |
368.429 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
3000 |
0.28 |
Functional ≤ 10μM
|
|
Z80566-2-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #2 Of 2), Other |
Other |
3000 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.91 |
17.91 |
-5.99 |
0 |
2 |
0 |
26 |
400.628 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VDR-1-E |
Vitamin D Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
VDR-1-E |
Vitamin D Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.42 |
Functional ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
8 |
0.38 |
Functional ≤ 10μM
|
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
50 |
0.34 |
Functional ≤ 10μM
|
|
Z80156-5-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #5 Of 12), Other |
Other |
3 |
0.40 |
Functional ≤ 10μM
|
|
Z80224-2-O |
MCF7 (Breast Carcinoma Cells) (cluster #2 Of 14), Other |
Other |
52 |
0.34 |
Functional ≤ 10μM
|
|
Z80437-1-O |
RWLeu4 (cluster #1 Of 1), Other |
Other |
6 |
0.38 |
Functional ≤ 10μM |
|
Z80437-1-O |
RWLeu4 (cluster #1 Of 1), Other |
Other |
6 |
0.38 |
Functional ≤ 10μM |
|
Z80566-2-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #2 Of 2), Other |
Other |
10 |
0.37 |
Functional ≤ 10μM |
|
Z80566-2-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #2 Of 2), Other |
Other |
17 |
0.36 |
Functional ≤ 10μM |
|
Z81117-2-O |
Keratinocytes (Keratinocytes) (cluster #2 Of 2), Other |
Other |
27 |
0.35 |
Functional ≤ 10μM
|
|
Z81298-1-O |
PE Cell Line (cluster #1 Of 1), Other |
Other |
100 |
0.33 |
Functional ≤ 10μM |
|
Z81338-2-O |
T-cells (cluster #2 Of 3), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.56 |
6.98 |
-5.55 |
3 |
3 |
0 |
61 |
416.646 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3S,5Z)-5-[(2E)-2-[(1S,3aS,7aR)-1-[(1R)-5-hydroxy-1,5-dimethyl-hexyl]-7a-methyl-2,3,3a,5,6,7-hexa
(1R,3S,5Z)-5-[(2E)-2-[(1S,3aS,7a…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80566-2-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #2 Of 2), Other |
Other |
17 |
0.36 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.56 |
6.86 |
-5.52 |
3 |
3 |
0 |
61 |
416.646 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80437-1-O |
RWLeu4 (cluster #1 Of 1), Other |
Other |
6 |
0.38 |
Functional ≤ 10μM |
|
Z80566-2-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #2 Of 2), Other |
Other |
17 |
0.36 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.56 |
7.04 |
-5.78 |
3 |
3 |
0 |
61 |
416.646 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80566-2-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #2 Of 2), Other |
Other |
17 |
0.36 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.56 |
7.09 |
-5.46 |
3 |
3 |
0 |
61 |
416.646 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
670 |
0.31 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
670 |
0.31 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
670 |
0.31 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
670 |
0.31 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.31 |
Functional ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.31 |
Functional ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.31 |
Functional ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.31 |
Functional ≤ 10μM
|
|
Z100709-1-O |
Squirrel Monkey (cluster #1 Of 1), Other |
Other |
600 |
0.31 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
8500 |
0.25 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
290 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.06 |
1.99 |
-9.25 |
0 |
3 |
0 |
31 |
373.496 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.06 |
2.1 |
-39.4 |
1 |
3 |
1 |
32 |
374.504 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.50 |
13.41 |
-10.21 |
2 |
3 |
0 |
49 |
395.587 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.23 |
-0.39 |
-9.74 |
1 |
5 |
0 |
60 |
381.259 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.62 |
14.36 |
-8.48 |
1 |
3 |
0 |
42 |
383.576 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.34 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
95 |
0.39 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
95 |
0.39 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
95 |
0.39 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
180 |
0.38 |
Binding ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
3467 |
0.31 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
4500 |
0.30 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
6120 |
0.29 |
Functional ≤ 10μM |
|
Z50592-1-O |
Oryctolagus Cuniculus (cluster #1 Of 8), Other |
Other |
800 |
0.34 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
1000 |
0.34 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
3630 |
0.30 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
10.45 |
-42.84 |
0 |
5 |
-1 |
82 |
342.415 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80566-2-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
9.53 |
-5.23 |
3 |
3 |
0 |
61 |
454.695 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.75 |
9.97 |
-67.37 |
1 |
9 |
-1 |
122 |
574.679 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
5.75 |
9.41 |
-24.1 |
2 |
9 |
0 |
119 |
575.687 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.69 |
12.06 |
-20.43 |
2 |
9 |
0 |
116 |
575.687 |
9 |
↓
|
|
|
|
Analogs
-
2000919
-
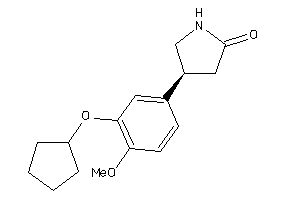
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
O77823-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3690 |
0.38 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
622 |
0.43 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
720 |
0.43 |
Binding ≤ 10μM
|
|
Q8VBU5-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
625 |
0.43 |
Functional ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 1), Other |
Other |
150 |
0.48 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
7 |
0.57 |
Binding ≤ 10μM |
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
880 |
0.42 |
Functional ≤ 10μM
|
|
Z100489-1-O |
Eosinophils (Eosinophils) (cluster #1 Of 1), Other |
Other |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
340 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
660 |
0.43 |
Functional ≤ 10μM |
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
4 |
0.59 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
40 |
0.52 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
1230 |
0.41 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
5.43 |
-11.26 |
1 |
4 |
0 |
48 |
275.348 |
4 |
↓
|
|
|
|
Analogs
-
4982
-
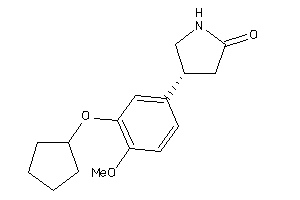
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
O77823-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
720 |
0.43 |
Binding ≤ 10μM
|
|
Q8VBU5-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
625 |
0.43 |
Functional ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 1), Other |
Other |
150 |
0.48 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
7 |
0.57 |
Binding ≤ 10μM |
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
880 |
0.42 |
Functional ≤ 10μM
|
|
Z100489-1-O |
Eosinophils (Eosinophils) (cluster #1 Of 1), Other |
Other |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
340 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
660 |
0.43 |
Functional ≤ 10μM |
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
4 |
0.59 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
40 |
0.52 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
1230 |
0.41 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
5.44 |
-11.23 |
1 |
4 |
0 |
48 |
275.348 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.85 |
2.11 |
-7.58 |
1 |
4 |
0 |
51 |
275.348 |
4 |
↓
|
|