|
|
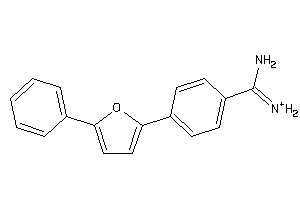
Analogs
-
1758871
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.38 |
Binding ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.38 |
Binding ≤ 10μM
|
|
Z50420-4-O |
Trypanosoma Brucei Brucei (cluster #4 Of 7), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
16 |
0.47 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
16 |
0.47 |
Functional ≤ 10μM
|
|
Z50459-1-O |
Leishmania Donovani (cluster #1 Of 8), Other |
Other |
6400 |
0.32 |
Functional ≤ 10μM
|
|
Z50466-7-O |
Trypanosoma Cruzi (cluster #7 Of 8), Other |
Other |
6400 |
0.32 |
Functional ≤ 10μM
|
|
Z50468-4-O |
Giardia Intestinalis (cluster #4 Of 4), Other |
Other |
440 |
0.39 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z50725-1-O |
Trypanosoma Brucei Rhodesiense (cluster #1 Of 7), Other |
Other |
4500 |
0.33 |
Functional ≤ 10μM
|
|
Z80035-5-O |
B16 (Melanoma Cells) (cluster #5 Of 7), Other |
Other |
9200 |
0.31 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
6400 |
0.32 |
Functional ≤ 10μM
|
|
Z81135-1-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #1 Of 6), Other |
Other |
6700 |
0.31 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
5.7 |
-76.45 |
8 |
5 |
2 |
116 |
306.369 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
8.65 |
-8.33 |
0 |
3 |
0 |
43 |
250.253 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
7.90 |
18.51 |
-6.65 |
0 |
4 |
0 |
68 |
536.797 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.39 |
-5.91 |
-77.64 |
9 |
7 |
2 |
139 |
283.339 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.93 |
-3.98 |
-78.76 |
8 |
5 |
2 |
116 |
280.331 |
3 |
↓
|
|
|
|
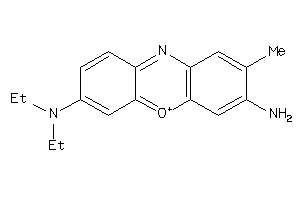
Analogs
-
4521067
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
11.31 |
-9.05 |
0 |
4 |
0 |
31 |
324.448 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
0.96 |
10.23 |
-19.87 |
0 |
4 |
1 |
32 |
324.448 |
5 |
↓
|
|
|
|
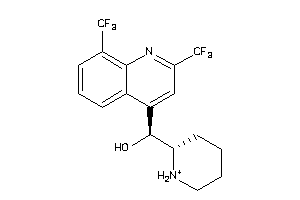
Analogs
-
3874185
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1221 |
0.32 |
Binding ≤ 10μM
|
|
AA2AR-2-E |
Adenosine A2a Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
124 |
0.37 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5623 |
0.28 |
Binding ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
47 |
0.39 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z50417-3-O |
Leishmania Infantum (cluster #3 Of 4), Other |
Other |
7000 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
91 |
0.38 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
7900 |
0.27 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 2), Other |
Other |
6500 |
0.28 |
ADME/T ≤ 10μM |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
4760 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
6.7 |
-52.23 |
3 |
3 |
1 |
50 |
379.324 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA2AR-2-E |
Adenosine A2a Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6553 |
0.28 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5250 |
0.28 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5623 |
0.28 |
Binding ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
47 |
0.39 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z50417-3-O |
Leishmania Infantum (cluster #3 Of 4), Other |
Other |
7000 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5 |
0.45 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
91 |
0.38 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
7900 |
0.27 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 2), Other |
Other |
6500 |
0.28 |
ADME/T ≤ 10μM |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
4760 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
6.35 |
-59.95 |
3 |
3 |
1 |
50 |
379.324 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.24 |
5.78 |
-11.48 |
2 |
3 |
0 |
45 |
378.316 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
255 |
0.36 |
Binding ≤ 10μM
|
|
AA2AR-2-E |
Adenosine A2a Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
61 |
0.39 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5623 |
0.28 |
Binding ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
47 |
0.39 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z50417-3-O |
Leishmania Infantum (cluster #3 Of 4), Other |
Other |
7000 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
91 |
0.38 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
7900 |
0.27 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 2), Other |
Other |
6500 |
0.28 |
ADME/T ≤ 10μM |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
4760 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
6.35 |
-59.95 |
3 |
3 |
1 |
50 |
379.324 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.24 |
5.69 |
-9.16 |
2 |
3 |
0 |
45 |
378.316 |
4 |
↓
|
|
|
|
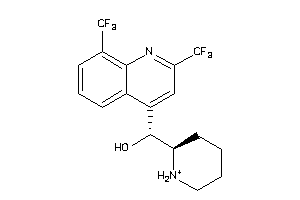
Analogs
-
537964
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8607 |
0.27 |
Binding ≤ 10μM
|
|
AA2AR-2-E |
Adenosine A2a Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2534 |
0.30 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5623 |
0.28 |
Binding ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
47 |
0.39 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z50417-3-O |
Leishmania Infantum (cluster #3 Of 4), Other |
Other |
7000 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
91 |
0.38 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
7900 |
0.27 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 2), Other |
Other |
6500 |
0.28 |
ADME/T ≤ 10μM |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
4760 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
6.7 |
-52.4 |
3 |
3 |
1 |
50 |
379.324 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
3 |
0.57 |
Functional ≤ 10μM
|
|
Z50725-7-O |
Trypanosoma Brucei Rhodesiense (cluster #7 Of 7), Other |
Other |
18 |
0.52 |
Functional ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
860 |
0.40 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.58 |
3.85 |
-76.3 |
9 |
5 |
2 |
119 |
279.347 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
-6.15 |
-84.43 |
8 |
6 |
2 |
121 |
472.181 |
8 |
↓
|
|
|
|
Analogs
-
6535287
-
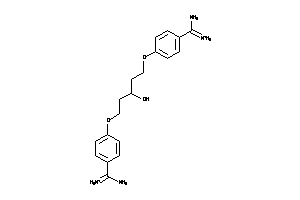
-
6535291
-
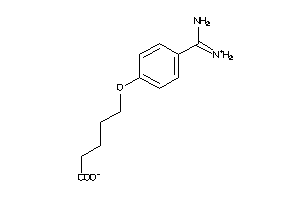
-
6535292
-
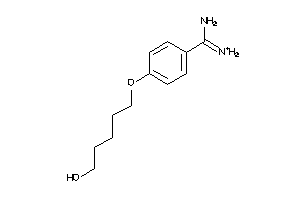
-
34928597
-
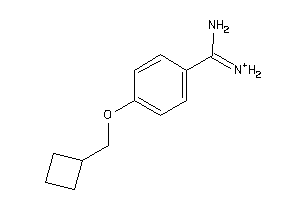
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S100B-2-E |
S-100 Protein Beta Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.34 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1160 |
0.33 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2300 |
0.32 |
Binding ≤ 10μM
|
|
Z104302-2-O |
Glutamate NMDA Receptor (cluster #2 Of 7), Other |
Other |
3060 |
0.31 |
Binding ≤ 10μM |
|
Z101865-1-O |
Leishmania Chagasi (cluster #1 Of 1), Other |
Other |
1110 |
0.33 |
Functional ≤ 10μM
|
|
Z101870-2-O |
Leishmania Guyanensis (cluster #2 Of 2), Other |
Other |
900 |
0.34 |
Functional ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
84 |
0.40 |
Functional ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
9100 |
0.28 |
Functional ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
6 |
0.46 |
Functional ≤ 10μM
|
|
Z50420-1-O |
Trypanosoma Brucei Brucei (cluster #1 Of 7), Other |
Other |
3 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
251 |
0.37 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
6 |
0.46 |
Functional ≤ 10μM
|
|
Z50457-1-O |
Leishmania Amazonensis (cluster #1 Of 3), Other |
Other |
460 |
0.35 |
Functional ≤ 10μM
|
|
Z50458-4-O |
Leishmania Braziliensis (cluster #4 Of 4), Other |
Other |
9800 |
0.28 |
Functional ≤ 10μM
|
|
Z50459-1-O |
Leishmania Donovani (cluster #1 Of 8), Other |
Other |
9800 |
0.28 |
Functional ≤ 10μM
|
|
Z50460-1-O |
Leishmania Major (cluster #1 Of 4), Other |
Other |
7500 |
0.29 |
Functional ≤ 10μM
|
|
Z50461-1-O |
Leishmania Mexicana (cluster #1 Of 3), Other |
Other |
9568 |
0.28 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
7100 |
0.29 |
Functional ≤ 10μM
|
|
Z50467-2-O |
Trichomonas Vaginalis (cluster #2 Of 3), Other |
Other |
3815 |
0.30 |
Functional ≤ 10μM
|
|
Z50468-4-O |
Giardia Intestinalis (cluster #4 Of 4), Other |
Other |
4079 |
0.30 |
Functional ≤ 10μM
|
|
Z50473-4-O |
Plasmodium Berghei (cluster #4 Of 5), Other |
Other |
2942 |
0.31 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
3 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
2500 |
0.31 |
Functional ≤ 10μM
|
|
Z50725-1-O |
Trypanosoma Brucei Rhodesiense (cluster #1 Of 7), Other |
Other |
50 |
0.41 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
40 |
0.41 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
2100 |
0.32 |
Functional ≤ 10μM
|
|
Z80178-2-O |
J774.A1 (Macrophage Cells) (cluster #2 Of 2), Other |
Other |
1030 |
0.34 |
ADME/T ≤ 10μM |
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 3), Other |
Other |
2700 |
0.31 |
ADME/T ≤ 10μM
|
|
Z81135-2-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #2 Of 6), Other |
Other |
6000 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.49 |
5.45 |
-74.02 |
8 |
6 |
2 |
122 |
342.443 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.68 |
1.99 |
-2.42 |
1 |
1 |
0 |
20 |
114.188 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.72 |
3.89 |
-75.76 |
8 |
6 |
2 |
122 |
314.389 |
8 |
↓
|
|
|
|
Analogs
-
7404548
-
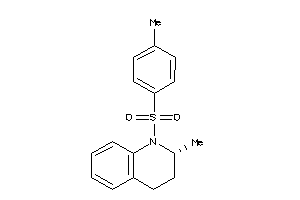
-
7404528
-
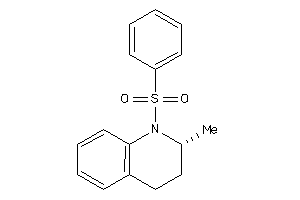
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
4900 |
0.35 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
8.06 |
-9.92 |
0 |
3 |
0 |
37 |
301.411 |
2 |
↓
|
|
|
|
Analogs
-
7404528
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
4900 |
0.35 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
8.38 |
-9.54 |
0 |
3 |
0 |
37 |
301.411 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
6650 |
0.35 |
Functional ≤ 10μM
|
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
7360 |
0.34 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.60 |
8.35 |
-7.98 |
0 |
3 |
0 |
37 |
366.28 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
6650 |
0.35 |
Functional ≤ 10μM
|
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
7360 |
0.34 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.60 |
8.02 |
-8.31 |
0 |
3 |
0 |
37 |
366.28 |
2 |
↓
|
|
|
|
Analogs
-
7404541
-
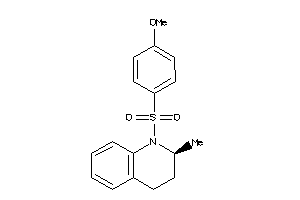
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
5990 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
7.02 |
-9.9 |
0 |
4 |
0 |
47 |
317.41 |
3 |
↓
|
|
|
|
Analogs
-
7404539
-
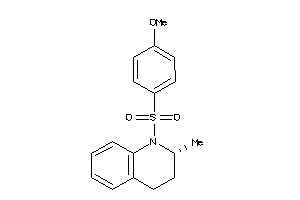
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
5990 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
6.69 |
-10.31 |
0 |
4 |
0 |
47 |
317.41 |
3 |
↓
|
|
|
|
Analogs
-
4098919
-
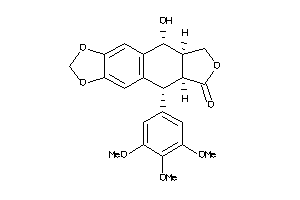
-
4098920
-
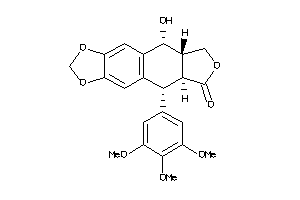
-
4475321
-
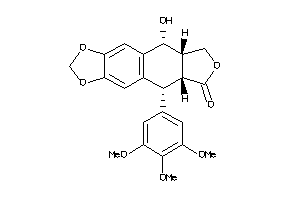
-
11682059
-
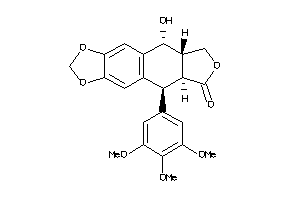
-
16969582
-
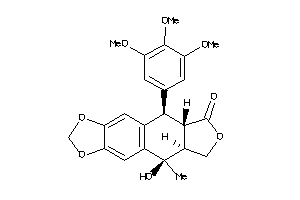
Draw
Identity
99%
90%
80%
70%
Vendors
And 52 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-1-E |
Glucocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.37 |
Binding ≤ 10μM
|
|
O46399-1-E |
Beta Tubulin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.26 |
Binding ≤ 10μM
|
|
Q862F3-2-E |
Tubulin Alpha Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM |
|
Q862L2-2-E |
Tubulin Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM |
|
Q9MZB0-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.26 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.27 |
Binding ≤ 10μM
|
|
TBB-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1560 |
0.27 |
Binding ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.30 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM |
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBG1-1-E |
Tubulin Gamma-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2180 |
0.26 |
Binding ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
590 |
0.29 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
590 |
0.29 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.30 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.30 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
590 |
0.29 |
Functional ≤ 10μM
|
|
Z100695-1-O |
WRL68 (Embryonic Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
4800 |
0.25 |
Functional ≤ 10μM
|
|
Z101973-1-O |
Paracentrotus Lividus (cluster #1 Of 1), Other |
Other |
500 |
0.29 |
Functional ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
10000 |
0.23 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
12 |
0.37 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
50 |
0.34 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
3 |
0.40 |
Functional ≤ 10μM
|
|
Z50651-2-O |
Vesicular Stomatitis Virus (cluster #2 Of 2), Other |
Other |
100 |
0.33 |
Functional ≤ 10μM
|
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.40 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
500 |
0.29 |
Functional ≤ 10μM
|
|
Z80110-1-O |
CV-1 (Kidney Cells) (cluster #1 Of 2), Other |
Other |
29 |
0.35 |
Functional ≤ 10μM |
|
Z80121-1-O |
DMS-79 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.36 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
52 |
0.34 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
2750 |
0.26 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
8100 |
0.24 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
50 |
0.34 |
Functional ≤ 10μM |
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
30 |
0.35 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
8500 |
0.24 |
Functional ≤ 10μM
|
|
Z80284-3-O |
MOLT-3 (T-lymphoblastic Leukemia Cells) (cluster #3 Of 3), Other |
Other |
14 |
0.37 |
Functional ≤ 10μM
|
|
Z80354-4-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
460 |
0.30 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
50 |
0.34 |
Functional ≤ 10μM |
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
13 |
0.37 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
18 |
0.36 |
Functional ≤ 10μM
|
|
Z80427-1-O |
RKO (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
29 |
0.35 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7630 |
0.24 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
9 |
0.38 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
13 |
0.37 |
Functional ≤ 10μM |
|
Z80847-1-O |
FaDu (Pharyngeal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
7 |
0.38 |
Functional ≤ 10μM
|
|
Z80866-1-O |
GLC4 Cell Line (cluster #1 Of 1), Other |
Other |
60 |
0.34 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
7 |
0.38 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
17 |
0.36 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
500 |
0.29 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
8 |
0.38 |
Functional ≤ 10μM
|
|
Z81135-1-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #1 Of 4), Other |
Other |
150 |
0.32 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
16 |
0.36 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
69 |
0.33 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
8 |
0.38 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
1000 |
0.28 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 1), Other |
Other |
150 |
0.32 |
ADME/T ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
34 |
0.35 |
ADME/T ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 3), Other |
Other |
23 |
0.36 |
ADME/T ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 3), Other |
Other |
5 |
0.39 |
ADME/T ≤ 10μM
|
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
9 |
0.38 |
ADME/T ≤ 10μM |
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 1), Other |
Other |
31 |
0.35 |
ADME/T ≤ 10μM
|
|
VE2-1-V |
Human Papillomavirus Regulatory Protein E2 (cluster #1 Of 1), Viral |
Viruses |
1050 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
4.95 |
-16.81 |
1 |
8 |
0 |
93 |
414.41 |
4 |
↓
|
|
|
|
Analogs
-
1530861
-
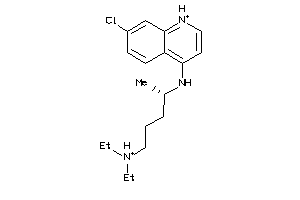
-
5850156
-
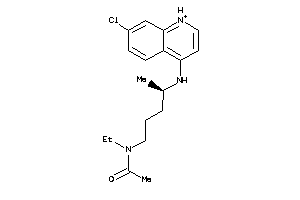
-
5850183
-
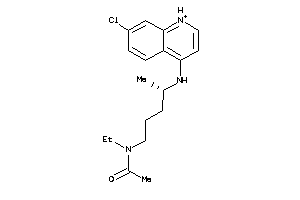
-
6424763
-
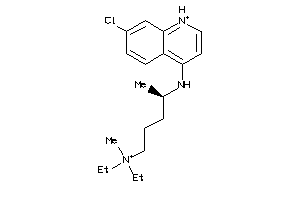
-
19144226
-
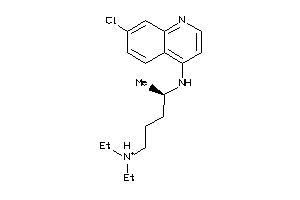
Draw
Identity
99%
90%
80%
70%
Vendors
And 66 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2512 |
0.36 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Binding ≤ 10μM
|
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
400 |
0.41 |
Binding ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100498-2-O |
Hepatocytes (cluster #2 Of 2), Other |
Other |
1900 |
0.36 |
Functional ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
1700 |
0.37 |
Functional ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
14 |
0.50 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z50136-2-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #2 Of 3), Other |
Other |
167 |
0.43 |
Functional ≤ 10μM
|
|
Z50424-2-O |
Cryptosporidium Parvum (cluster #2 Of 2), Other |
Other |
27 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
900 |
0.38 |
Functional ≤ 10μM
|
|
Z50468-2-O |
Giardia Intestinalis (cluster #2 Of 4), Other |
Other |
401 |
0.41 |
Functional ≤ 10μM
|
|
Z50473-4-O |
Plasmodium Berghei (cluster #4 Of 5), Other |
Other |
72 |
0.45 |
Functional ≤ 10μM
|
|
Z50474-1-O |
Plasmodium Yoelii (cluster #1 Of 1), Other |
Other |
106 |
0.44 |
Functional ≤ 10μM
|
|
Z80136-1-O |
FM3A (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
40 |
0.47 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
4000 |
0.34 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
9670 |
0.32 |
Functional ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 2), Other |
Other |
5100 |
0.34 |
ADME/T ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
32 |
0.48 |
ADME/T ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
64 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
8300 |
0.32 |
ADME/T ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
600 |
0.40 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
7700 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
10.55 |
-81.59 |
3 |
3 |
2 |
31 |
321.896 |
8 |
↓
|
|
|
|
Analogs
-
5850156
-

-
5850183
-

-
6424763
-

-
19144226
-

-
19144231
-
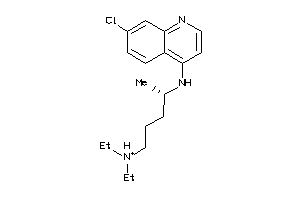
Draw
Identity
99%
90%
80%
70%
Vendors
And 64 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2512 |
0.36 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Binding ≤ 10μM
|
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
400 |
0.41 |
Binding ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100498-2-O |
Hepatocytes (cluster #2 Of 2), Other |
Other |
1900 |
0.36 |
Functional ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
1700 |
0.37 |
Functional ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
14 |
0.50 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z50136-2-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #2 Of 3), Other |
Other |
167 |
0.43 |
Functional ≤ 10μM
|
|
Z50424-2-O |
Cryptosporidium Parvum (cluster #2 Of 2), Other |
Other |
27 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
900 |
0.38 |
Functional ≤ 10μM
|
|
Z50468-2-O |
Giardia Intestinalis (cluster #2 Of 4), Other |
Other |
401 |
0.41 |
Functional ≤ 10μM
|
|
Z50473-4-O |
Plasmodium Berghei (cluster #4 Of 5), Other |
Other |
72 |
0.45 |
Functional ≤ 10μM
|
|
Z50474-1-O |
Plasmodium Yoelii (cluster #1 Of 1), Other |
Other |
106 |
0.44 |
Functional ≤ 10μM
|
|
Z80136-1-O |
FM3A (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
40 |
0.47 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
4000 |
0.34 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
9670 |
0.32 |
Functional ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 2), Other |
Other |
5100 |
0.34 |
ADME/T ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
32 |
0.48 |
ADME/T ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
64 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
8300 |
0.32 |
ADME/T ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
600 |
0.40 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
7700 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
10.55 |
-81.48 |
3 |
3 |
2 |
31 |
321.896 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.77 |
-5.22 |
-71.17 |
10 |
6 |
2 |
127 |
284.367 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-1-O |
Trypanosoma Brucei (cluster #1 Of 6), Other |
Other |
3000 |
0.43 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
755 |
0.48 |
Functional ≤ 10μM |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50459-8-O |
Leishmania Donovani (cluster #8 Of 8), Other |
Other |
2680 |
0.43 |
Functional ≤ 10μM
|
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
220 |
0.52 |
Functional ≤ 10μM
|
|
Z50725-7-O |
Trypanosoma Brucei Rhodesiense (cluster #7 Of 7), Other |
Other |
600 |
0.48 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
1500 |
0.45 |
Functional ≤ 10μM
|
|
Z80695-2-O |
BE (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
1280 |
0.46 |
Functional ≤ 10μM
|
|
Z81240-1-O |
MAC15A (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
3950 |
0.42 |
Functional ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
1050 |
0.47 |
ADME/T ≤ 10μM |
|
Z81057-4-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #4 Of 5), Other |
Other |
1180 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
1120 |
0.46 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
0.21 |
-16.41 |
0 |
2 |
0 |
17 |
232.286 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 53 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
832 |
0.34 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Binding ≤ 10μM
|
|
PPARD-2-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3620 |
0.30 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.44 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.40 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
325 |
0.36 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.40 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
210 |
0.37 |
Functional ≤ 10μM
|
|
Z80106-1-O |
COS-1 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.43 |
Functional ≤ 10μM
|
|
Z80169-1-O |
Huh-7 (Hepatocellular Carcinoma) (cluster #1 Of 1), Other |
Other |
220 |
0.37 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z81117-1-O |
Keratinocytes (Keratinocytes) (cluster #1 Of 2), Other |
Other |
8000 |
0.29 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
5000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
7.4 |
-33.54 |
2 |
6 |
1 |
73 |
358.443 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.35 |
7.28 |
-11.07 |
1 |
6 |
0 |
72 |
357.435 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 54 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
832 |
0.34 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Binding ≤ 10μM
|
|
PPARD-2-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3620 |
0.30 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.44 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.41 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
325 |
0.36 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.40 |
Functional ≤ 10μM
|
|
Z80106-1-O |
COS-1 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.43 |
Functional ≤ 10μM
|
|
Z80169-1-O |
Huh-7 (Hepatocellular Carcinoma) (cluster #1 Of 1), Other |
Other |
220 |
0.37 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z81117-1-O |
Keratinocytes (Keratinocytes) (cluster #1 Of 2), Other |
Other |
8000 |
0.29 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
5000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
7.4 |
-33.23 |
2 |
6 |
1 |
73 |
358.443 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.35 |
7.28 |
-11.54 |
1 |
6 |
0 |
72 |
357.435 |
7 |
↓
|
|
|
|
Analogs
-
8101062
-
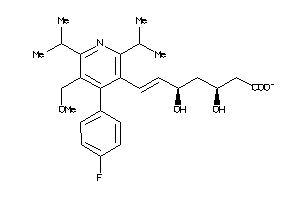
-
9831786
-
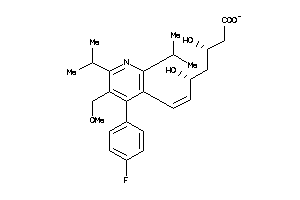
-
9831787
-
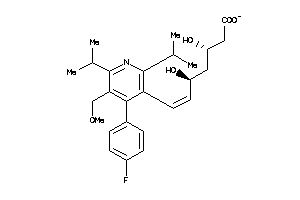
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-2-E |
HMG-CoA Reductase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
Z81135-1-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #1 Of 4), Other |
Other |
7 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
9.79 |
-51.62 |
2 |
6 |
-1 |
103 |
458.55 |
11 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.45 |
10.19 |
-65.02 |
3 |
6 |
0 |
104 |
459.558 |
11 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
340 |
0.50 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
1300 |
0.46 |
Functional ≤ 10μM
|
|
Z80120-4-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #4 Of 5), Other |
Other |
1440 |
0.45 |
Functional ≤ 10μM
|
|
Z80682-11-O |
A549 (Lung Carcinoma Cells) (cluster #11 Of 11), Other |
Other |
550 |
0.49 |
Functional ≤ 10μM
|
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 4), Other |
Other |
8100 |
0.40 |
Functional ≤ 10μM
|
|
Z81225-2-O |
M109 (Lung Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
1000 |
0.47 |
Functional ≤ 10μM
|
|
Z81240-1-O |
MAC15A (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
9650 |
0.39 |
Functional ≤ 10μM
|
|
Z100094-1-O |
MLF (Lung Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
4900 |
0.41 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.42 |
8.58 |
-23.57 |
1 |
2 |
1 |
20 |
233.294 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPARD-2-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.34 |
Binding ≤ 10μM
|
|
PPARD-1-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.30 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
30 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.35 |
0.11 |
-48.73 |
0 |
3 |
-1 |
49 |
567.706 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
990 |
0.28 |
Binding ≤ 10μM
|
|
PPARD-2-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.38 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
990 |
0.28 |
Functional ≤ 10μM
|
|
PPARD-1-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
85 |
0.33 |
Functional ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.28 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
6 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.85 |
1.99 |
-48.81 |
0 |
4 |
-1 |
62 |
452.499 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABPI-3-E |
Fatty Acid Binding Protein Intestinal (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
FABPL-3-E |
Fatty Acid-binding Protein, Liver (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
94 |
0.45 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4500 |
0.34 |
Binding ≤ 10μM |
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9200 |
0.32 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
3000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
1.72 |
-50.04 |
0 |
4 |
-1 |
66 |
317.748 |
5 |
↓
|
|