|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100771-2-O |
Mitochondrial Complex I (cluster #2 Of 2), Other |
Other |
5 |
0.39 |
Binding ≤ 10μM
|
|
Z81137-1-O |
L929 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.83 |
7.55 |
-9.74 |
2 |
5 |
0 |
72 |
415.574 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100732-1-O |
CNE (Nasopharyngeal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
700 |
0.41 |
Functional ≤ 10μM |
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
440 |
0.42 |
Functional ≤ 10μM
|
|
Z80186-4-O |
K562 (Erythroleukemia Cells) (cluster #4 Of 11), Other |
Other |
630 |
0.41 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1900 |
0.38 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
230 |
0.44 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
240 |
0.44 |
Functional ≤ 10μM
|
|
Z81252-1-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #1 Of 11), Other |
Other |
6600 |
0.35 |
Functional ≤ 10μM |
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
7300 |
0.34 |
Functional ≤ 10μM |
|
Z81137-1-O |
L929 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
3200 |
0.37 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
3.58 |
-8.99 |
3 |
5 |
0 |
95 |
288.299 |
3 |
↓
|
|
|
|
Analogs
-
116934
-
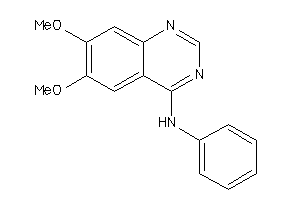
-
3815316
-
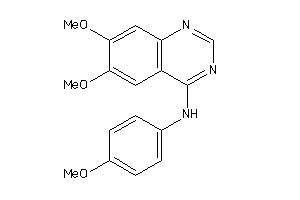
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.55 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Binding ≤ 10μM |
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.35 |
Binding ≤ 10μM |
|
Z81137-2-O |
L929 (Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
1700 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
-1.4 |
-11.31 |
2 |
6 |
0 |
76 |
297.314 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
1529898
-
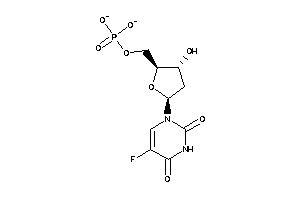
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-5.84 |
-140.72 |
2 |
10 |
-2 |
156 |
324.157 |
4 |
↓
|
|
|
|
Analogs
-
1529898
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-4.64 |
-163.61 |
2 |
10 |
-2 |
156 |
324.157 |
4 |
↓
|
|
|
|
Analogs
-
1529898
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-5.54 |
-139.88 |
2 |
10 |
-2 |
156 |
324.157 |
4 |
↓
|
|
|
|
Analogs
-
388678
-
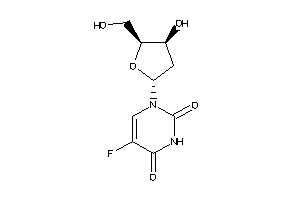
-
489135
-
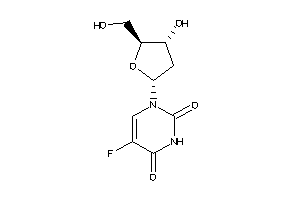
-
3813010
-
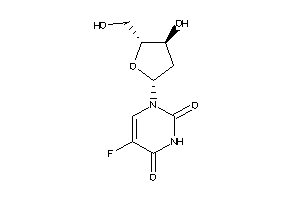
-
3830846
-
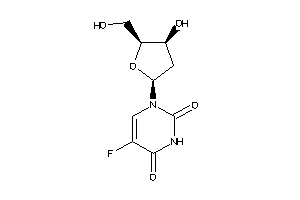
-
3977743
-
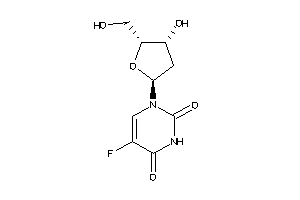
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.42 |
Binding ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
500 |
0.52 |
Functional ≤ 10μM
|
|
Z80101-2-O |
COLO 320DM (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
650 |
0.51 |
Functional ≤ 10μM |
|
Z80120-3-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
92 |
0.58 |
Functional ≤ 10μM |
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
45 |
0.61 |
Functional ≤ 10μM
|
|
Z80208-1-O |
L5178Y (Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
2 |
0.72 |
Functional ≤ 10μM
|
|
Z80268-2-O |
MKN-28 (Gastric Epithelial Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80269-3-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
3500 |
0.45 |
Functional ≤ 10μM |
|
Z80347-2-O |
NUGC-3 (Gastric Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
15 |
0.64 |
Functional ≤ 10μM |
|
Z80525-2-O |
SW48 (cluster #2 Of 2), Other |
Other |
130 |
0.57 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM |
|
Z80686-1-O |
AZ-521 Cell Line (cluster #1 Of 1), Other |
Other |
50 |
0.60 |
Functional ≤ 10μM |
|
Z81048-1-O |
TSU (Prostatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
58 |
0.60 |
Functional ≤ 10μM
|
|
Z81121-1-O |
KKLS Cell Line (cluster #1 Of 1), Other |
Other |
760 |
0.50 |
Functional ≤ 10μM |
|
Z81137-1-O |
L929 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
7700 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-4-O |
LNCaP (Prostate Carcinoma) (cluster #4 Of 5), Other |
Other |
69 |
0.59 |
Functional ≤ 10μM
|
|
Z81330-1-O |
SNU- (cluster #1 Of 1), Other |
Other |
3 |
0.70 |
Functional ≤ 10μM |
|
Z81335-3-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
49 |
0.60 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.72 |
-5.12 |
-11.33 |
3 |
7 |
0 |
105 |
246.194 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.26 |
-7.87 |
-48.56 |
2 |
7 |
-1 |
108 |
245.186 |
2 |
↓
|
|
|
|
Analogs
-
3977743
-

-
3977744
-
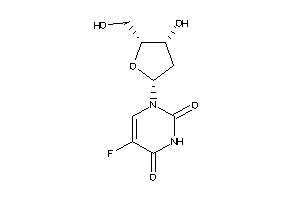
-
4045293
-
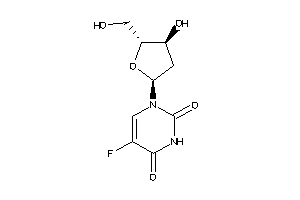
-
1457
-
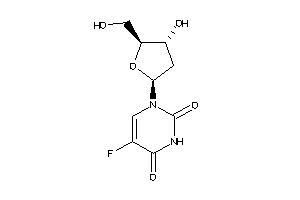
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.42 |
Binding ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
500 |
0.52 |
Functional ≤ 10μM
|
|
Z80101-2-O |
COLO 320DM (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
650 |
0.51 |
Functional ≤ 10μM |
|
Z80120-3-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
92 |
0.58 |
Functional ≤ 10μM |
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
45 |
0.61 |
Functional ≤ 10μM
|
|
Z80208-1-O |
L5178Y (Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
2 |
0.72 |
Functional ≤ 10μM
|
|
Z80268-2-O |
MKN-28 (Gastric Epithelial Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80269-3-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
3500 |
0.45 |
Functional ≤ 10μM |
|
Z80347-2-O |
NUGC-3 (Gastric Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
15 |
0.64 |
Functional ≤ 10μM |
|
Z80525-2-O |
SW48 (cluster #2 Of 2), Other |
Other |
130 |
0.57 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM |
|
Z80686-1-O |
AZ-521 Cell Line (cluster #1 Of 1), Other |
Other |
50 |
0.60 |
Functional ≤ 10μM |
|
Z81048-1-O |
TSU (Prostatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
58 |
0.60 |
Functional ≤ 10μM
|
|
Z81121-1-O |
KKLS Cell Line (cluster #1 Of 1), Other |
Other |
760 |
0.50 |
Functional ≤ 10μM |
|
Z81137-1-O |
L929 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
7700 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-4-O |
LNCaP (Prostate Carcinoma) (cluster #4 Of 5), Other |
Other |
69 |
0.59 |
Functional ≤ 10μM
|
|
Z81330-1-O |
SNU- (cluster #1 Of 1), Other |
Other |
3 |
0.70 |
Functional ≤ 10μM |
|
Z81335-3-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
49 |
0.60 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.72 |
-4.03 |
-17.41 |
3 |
7 |
0 |
105 |
246.194 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.26 |
-6.78 |
-57.51 |
2 |
7 |
-1 |
108 |
245.186 |
2 |
↓
|
|
|
|
Analogs
-
3977743
-

-
3977744
-

-
4045293
-

-
1457
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.42 |
Binding ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
500 |
0.52 |
Functional ≤ 10μM
|
|
Z80101-2-O |
COLO 320DM (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
650 |
0.51 |
Functional ≤ 10μM |
|
Z80120-3-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
92 |
0.58 |
Functional ≤ 10μM |
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
45 |
0.61 |
Functional ≤ 10μM
|
|
Z80208-1-O |
L5178Y (Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
2 |
0.72 |
Functional ≤ 10μM
|
|
Z80268-2-O |
MKN-28 (Gastric Epithelial Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80269-3-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
3500 |
0.45 |
Functional ≤ 10μM |
|
Z80347-2-O |
NUGC-3 (Gastric Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
15 |
0.64 |
Functional ≤ 10μM |
|
Z80525-2-O |
SW48 (cluster #2 Of 2), Other |
Other |
130 |
0.57 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM |
|
Z80686-1-O |
AZ-521 Cell Line (cluster #1 Of 1), Other |
Other |
50 |
0.60 |
Functional ≤ 10μM |
|
Z81048-1-O |
TSU (Prostatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
58 |
0.60 |
Functional ≤ 10μM
|
|
Z81121-1-O |
KKLS Cell Line (cluster #1 Of 1), Other |
Other |
760 |
0.50 |
Functional ≤ 10μM |
|
Z81137-1-O |
L929 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
7700 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-4-O |
LNCaP (Prostate Carcinoma) (cluster #4 Of 5), Other |
Other |
69 |
0.59 |
Functional ≤ 10μM
|
|
Z81330-1-O |
SNU- (cluster #1 Of 1), Other |
Other |
3 |
0.70 |
Functional ≤ 10μM |
|
Z81335-3-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
49 |
0.60 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.72 |
-4.23 |
-22.26 |
3 |
7 |
0 |
105 |
246.194 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.26 |
-6.98 |
-63.95 |
2 |
7 |
-1 |
108 |
245.186 |
2 |
↓
|
|
|
|
Analogs
-
3977743
-

-
3977744
-

-
4045293
-

-
1457
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.42 |
Binding ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
500 |
0.52 |
Functional ≤ 10μM
|
|
Z80101-2-O |
COLO 320DM (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
650 |
0.51 |
Functional ≤ 10μM |
|
Z80120-3-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
92 |
0.58 |
Functional ≤ 10μM |
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
45 |
0.61 |
Functional ≤ 10μM
|
|
Z80208-1-O |
L5178Y (Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
2 |
0.72 |
Functional ≤ 10μM
|
|
Z80268-2-O |
MKN-28 (Gastric Epithelial Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80269-3-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
3500 |
0.45 |
Functional ≤ 10μM |
|
Z80347-2-O |
NUGC-3 (Gastric Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
15 |
0.64 |
Functional ≤ 10μM |
|
Z80525-2-O |
SW48 (cluster #2 Of 2), Other |
Other |
130 |
0.57 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM |
|
Z80686-1-O |
AZ-521 Cell Line (cluster #1 Of 1), Other |
Other |
50 |
0.60 |
Functional ≤ 10μM |
|
Z81048-1-O |
TSU (Prostatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
58 |
0.60 |
Functional ≤ 10μM
|
|
Z81121-1-O |
KKLS Cell Line (cluster #1 Of 1), Other |
Other |
760 |
0.50 |
Functional ≤ 10μM |
|
Z81137-1-O |
L929 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
7700 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-4-O |
LNCaP (Prostate Carcinoma) (cluster #4 Of 5), Other |
Other |
69 |
0.59 |
Functional ≤ 10μM
|
|
Z81330-1-O |
SNU- (cluster #1 Of 1), Other |
Other |
3 |
0.70 |
Functional ≤ 10μM |
|
Z81335-3-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
49 |
0.60 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.72 |
-4.89 |
-10.74 |
3 |
7 |
0 |
105 |
246.194 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.26 |
-7.64 |
-47.75 |
2 |
7 |
-1 |
108 |
245.186 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
4098238
-
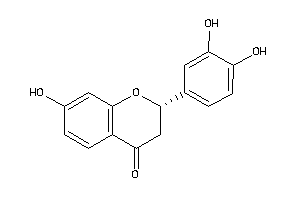
-
39091
-
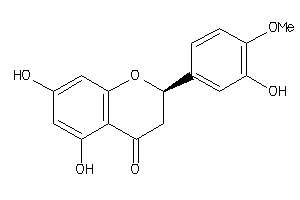
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1284 |
0.39 |
Binding ≤ 10μM
|
|
Z81137-1-O |
L929 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
6000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.63 |
-1.2 |
-12.56 |
4 |
6 |
0 |
107 |
288.255 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.63 |
-0.21 |
-53.53 |
3 |
6 |
-1 |
110 |
287.247 |
1 |
↓
|
|