|
|
Analogs
-
34380715
-
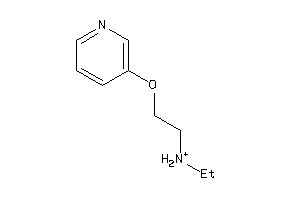
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
Z104283-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #2 Of 4), Other |
Other |
35 |
0.95 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.71 |
2.31 |
-40.34 |
2 |
3 |
1 |
39 |
153.205 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.71 |
2.59 |
-88.63 |
3 |
3 |
2 |
40 |
154.213 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA2-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA5-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA6-3-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA9-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHB2-6-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHB3-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHB4-6-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
8.25 |
-39.49 |
3 |
3 |
1 |
45 |
340.487 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3540 |
0.48 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
322 |
0.57 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3540 |
0.48 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3540 |
0.48 |
Binding ≤ 10μM
|
|
ACHG-1-E |
Acetylcholine Receptor Protein Gamma Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3540 |
0.48 |
Binding ≤ 10μM
|
|
Z104282-1-O |
Acetylcholine Receptor; Alpha1/beta1/delta/gamma (cluster #1 Of 1), Other |
Other |
8200 |
0.45 |
Binding ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
617 |
0.54 |
Binding ≤ 10μM
|
|
Z104288-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 2), Other |
Other |
240 |
0.58 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
-2.77 |
-43.65 |
2 |
3 |
1 |
42 |
212.276 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.37 |
14.31 |
-14.39 |
0 |
6 |
0 |
76 |
526.592 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
43 |
0.86 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
2.92 |
-40.51 |
2 |
3 |
1 |
39 |
167.232 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.76 |
3.17 |
-87.11 |
3 |
3 |
2 |
40 |
168.24 |
4 |
↓
|
|
|
|
Analogs
-
3705296
-
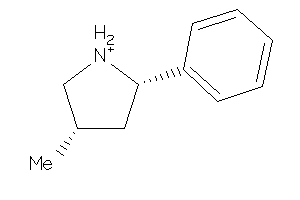
-
3705297
-
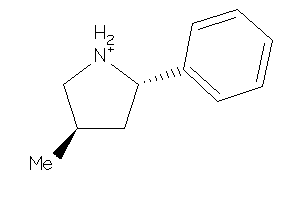
-
3705298
-
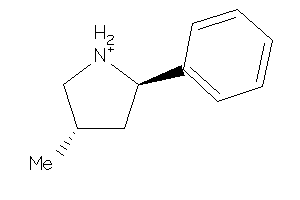
-
3705299
-
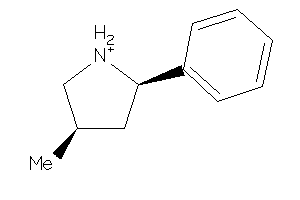
Draw
Identity
99%
90%
80%
70%
Vendors
And 74 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
5.45 |
-35.2 |
2 |
1 |
1 |
17 |
148.229 |
1 |
↓
|
|
|
|
Analogs
-
3705296
-

-
3705297
-

-
3705298
-

-
3705299
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 73 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
8696 |
0.64 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
5.45 |
-35.49 |
2 |
1 |
1 |
17 |
148.229 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
8100 |
0.45 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
132 |
0.60 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
2100 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
8.01 |
-38.99 |
1 |
2 |
1 |
17 |
213.304 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
60 |
0.34 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
7810 |
0.24 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
100 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.09 |
3.5 |
-63.39 |
0 |
2 |
2 |
7 |
398.594 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
27 |
0.88 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.34 |
0.17 |
-40.61 |
4 |
4 |
1 |
65 |
168.22 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.34 |
0.45 |
-87.59 |
5 |
4 |
2 |
66 |
169.228 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
35 |
0.55 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
3600 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.13 |
4.38 |
-50.34 |
3 |
5 |
1 |
70 |
262.333 |
3 |
↓
|
|
|
|
Analogs
-
6528452
-
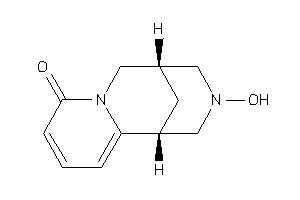
Draw
Identity
99%
90%
80%
70%
Vendors
And 46 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
250 |
0.66 |
Binding ≤ 10μM
|
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.82 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
144 |
0.68 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
900 |
0.60 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5820 |
0.52 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
250 |
0.66 |
Binding ≤ 10μM
|
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2700 |
0.56 |
Binding ≤ 10μM
|
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
900 |
0.60 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.66 |
Binding ≤ 10μM
|
|
ACHG-1-E |
Acetylcholine Receptor Protein Gamma Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.66 |
Binding ≤ 10μM
|
|
Z104281-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta2 (cluster #2 Of 2), Other |
Other |
37 |
0.74 |
Binding ≤ 10μM
|
|
Z104282-1-O |
Acetylcholine Receptor; Alpha1/beta1/delta/gamma (cluster #1 Of 1), Other |
Other |
250 |
0.66 |
Binding ≤ 10μM
|
|
Z104283-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #2 Of 4), Other |
Other |
3 |
0.85 |
Binding ≤ 10μM
|
|
Z104284-1-O |
Neuronal Acetylcholine Receptor; Alpha2/beta4 (cluster #1 Of 1), Other |
Other |
5 |
0.83 |
Binding ≤ 10μM
|
|
Z104286-2-O |
Neuronal Acetylcholine Receptor; Alpha2/beta2 (cluster #2 Of 2), Other |
Other |
1 |
0.90 |
Binding ≤ 10μM
|
|
Z104287-3-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #3 Of 3), Other |
Other |
217 |
0.67 |
Binding ≤ 10μM
|
|
Z104288-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 2), Other |
Other |
840 |
0.61 |
Binding ≤ 10μM
|
|
Z104289-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #2 Of 2), Other |
Other |
2 |
0.87 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
69 |
0.72 |
Binding ≤ 10μM
|
|
Z104282-1-O |
Acetylcholine Receptor; Alpha1/beta1/delta/gamma (cluster #1 Of 1), Other |
Other |
11 |
0.80 |
Functional ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z104287-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #2 Of 2), Other |
Other |
25 |
0.76 |
Functional ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 1), Other |
Other |
9 |
0.80 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
60 |
0.72 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.27 |
4.62 |
-47.94 |
2 |
3 |
1 |
39 |
191.254 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.69 |
14.27 |
-20.96 |
0 |
4 |
0 |
57 |
378.468 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHA2-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHA5-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHA6-3-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHA9-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHB2-6-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHB3-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
|
ACHB4-6-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.76 |
7.06 |
-40.49 |
1 |
2 |
1 |
22 |
178.255 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
11 |
0.53 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4260 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
9.27 |
-45.82 |
1 |
4 |
1 |
33 |
281.383 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.02 |
9.36 |
-88.52 |
2 |
4 |
2 |
35 |
282.391 |
2 |
↓
|
|
|
|
Analogs
-
53131873
-
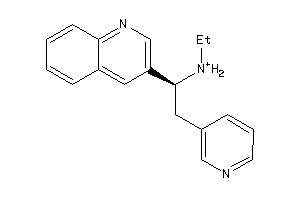
-
53131878
-
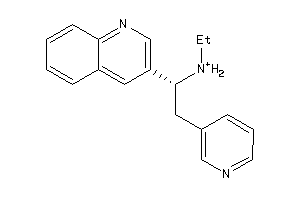
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
5.81 |
-42.9 |
2 |
2 |
1 |
29 |
199.277 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.07 |
6.1 |
-99.3 |
3 |
2 |
2 |
31 |
200.285 |
1 |
↓
|
|
|
|
Analogs
-
53131873
-

-
53131878
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
93 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
5.81 |
-42.86 |
2 |
2 |
1 |
29 |
199.277 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.07 |
6.1 |
-99.36 |
3 |
2 |
2 |
31 |
200.285 |
1 |
↓
|
|