|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
17 |
0.91 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
2 |
1.01 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
17 |
0.91 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
17 |
0.91 |
Binding ≤ 10μM
|
|
ACHD-3-E |
Acetylcholine Receptor Protein Delta Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.72 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.72 |
Functional ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8900 |
0.59 |
Functional ≤ 10μM
|
|
Z104281-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta2 (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104283-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #2 Of 4), Other |
Other |
700 |
0.72 |
Binding ≤ 10μM
|
|
Z104286-2-O |
Neuronal Acetylcholine Receptor; Alpha2/beta2 (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104287-3-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #3 Of 3), Other |
Other |
72 |
0.83 |
Binding ≤ 10μM
|
|
Z104287-3-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #3 Of 3), Other |
Other |
74 |
0.83 |
Binding ≤ 10μM
|
|
Z104289-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #2 Of 2), Other |
Other |
7 |
0.95 |
Binding ≤ 10μM
|
|
Z104290-4-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #4 Of 4), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104290-4-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #4 Of 4), Other |
Other |
2 |
1.01 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3 |
0.99 |
Functional ≤ 10μM
|
|
Z80175-1-O |
IMR-32 (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
700 |
0.72 |
Functional ≤ 10μM
|
|
Z81074-1-O |
K177cell Line (cluster #1 Of 1), Other |
Other |
700 |
0.72 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.83 |
-2.56 |
-40.53 |
2 |
3 |
1 |
38 |
165.216 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.83 |
-2.45 |
-88.84 |
3 |
3 |
2 |
39 |
166.224 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.81 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1530 |
0.58 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
8 |
0.81 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
ACHD-3-E |
Acetylcholine Receptor Protein Delta Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104283-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #2 Of 4), Other |
Other |
750 |
0.61 |
Binding ≤ 10μM
|
|
Z104287-3-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #3 Of 3), Other |
Other |
1400 |
0.59 |
Binding ≤ 10μM
|
|
Z104290-4-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #4 Of 4), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104290-4-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #4 Of 4), Other |
Other |
2 |
0.87 |
Binding ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 2), Other |
Other |
750 |
0.61 |
Functional ≤ 10μM |
|
Z81074-1-O |
K177cell Line (cluster #1 Of 1), Other |
Other |
750 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
0.08 |
-37.62 |
1 |
3 |
1 |
26 |
193.27 |
3 |
↓
|
|
|
|
Analogs
-
26717002
-
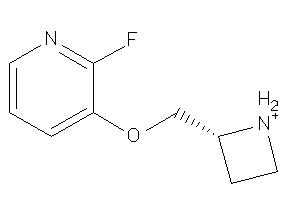
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
181 |
0.73 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
3680 |
0.59 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
188 |
0.72 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACHD-3-E |
Acetylcholine Receptor Protein Delta Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104290-4-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #4 Of 4), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.11 |
-1.94 |
-44.73 |
2 |
3 |
1 |
38 |
183.206 |
3 |
↓
|
|
|
|
Analogs
-
6525315
-
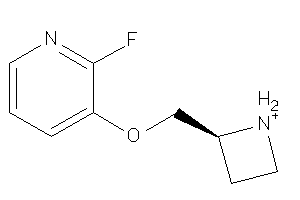
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHD-3-E |
Acetylcholine Receptor Protein Delta Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104290-4-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #4 Of 4), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
2.18 |
-44.24 |
2 |
3 |
1 |
39 |
183.206 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHD-3-E |
Acetylcholine Receptor Protein Delta Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104290-4-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #4 Of 4), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
3.43 |
-40.62 |
2 |
3 |
1 |
39 |
291.112 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHD-3-E |
Acetylcholine Receptor Protein Delta Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
-3.4 |
-41.59 |
2 |
3 |
1 |
38 |
291.112 |
3 |
↓
|
|
|
|
Analogs
-
6562
-
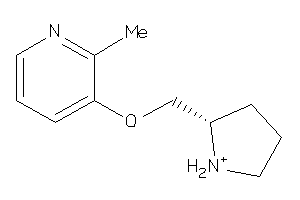
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHD-3-E |
Acetylcholine Receptor Protein Delta Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
39 |
0.74 |
Binding ≤ 10μM
|
|
Z104290-4-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #4 Of 4), Other |
Other |
39 |
0.74 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
3.95 |
-39.66 |
2 |
3 |
1 |
39 |
193.27 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.32 |
4.22 |
-85.01 |
3 |
3 |
2 |
40 |
194.278 |
3 |
↓
|
|
|
|
Analogs
-
18379
-
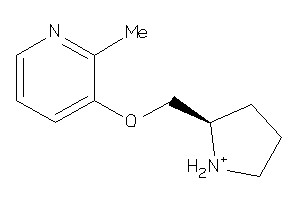
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHD-3-E |
Acetylcholine Receptor Protein Delta Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
17 |
0.78 |
Binding ≤ 10μM
|
|
Z104290-4-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #4 Of 4), Other |
Other |
19 |
0.77 |
Binding ≤ 10μM
|
|
Z104290-4-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #4 Of 4), Other |
Other |
39 |
0.74 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
1100 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
4 |
-39.9 |
2 |
3 |
1 |
39 |
193.27 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.32 |
4.26 |
-84.98 |
3 |
3 |
2 |
40 |
194.278 |
3 |
↓
|
|
|
|
Analogs
-
13704064
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHD-3-E |
Acetylcholine Receptor Protein Delta Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z80175-1-O |
IMR-32 (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
5800 |
0.56 |
Functional ≤ 10μM
|
|
Z81074-1-O |
K177cell Line (cluster #1 Of 1), Other |
Other |
4300 |
0.58 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
3.22 |
-37.97 |
2 |
3 |
1 |
39 |
179.243 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.10 |
3.5 |
-87.79 |
3 |
3 |
2 |
40 |
180.251 |
3 |
↓
|
|