|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.26 |
2.61 |
-61.64 |
0 |
7 |
-1 |
87 |
304.307 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 33 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A0ZX81-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
80 |
0.50 |
Binding ≤ 10μM
|
|
A1E3K9-1-B |
Beta-lactamase SCO-1 (cluster #1 Of 1), Bacterial |
Bacteria |
10 |
0.56 |
Binding ≤ 10μM
|
|
A2RP81-1-B |
PER-2 Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
96 |
0.49 |
Binding ≤ 10μM
|
|
A4KCT8-1-B |
Gil1 (cluster #1 Of 1), Bacterial |
Bacteria |
40 |
0.52 |
Binding ≤ 10μM
|
|
A8RR46-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
4 |
0.59 |
Binding ≤ 10μM
|
|
AMPC-5-B |
Beta-lactamase (cluster #5 Of 6), Bacterial |
Bacteria |
190 |
0.47 |
Binding ≤ 10μM
|
|
B8R6A5-1-B |
PenB Class A Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
500 |
0.44 |
Binding ≤ 10μM
|
|
BLA1-1-B |
Beta-lactamase L1 (cluster #1 Of 3), Bacterial |
Bacteria |
110 |
0.49 |
Binding ≤ 10μM
|
|
BLAC-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
290 |
0.46 |
Binding ≤ 10μM
|
|
BLAC-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
297 |
0.46 |
Binding ≤ 10μM
|
|
BLAC-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
510 |
0.44 |
Binding ≤ 10μM
|
|
BLAT-3-B |
Beta-lactamase TEM (cluster #3 Of 4), Bacterial |
Bacteria |
60 |
0.51 |
Binding ≤ 10μM
|
|
BLAT-3-B |
Beta-lactamase TEM (cluster #3 Of 4), Bacterial |
Bacteria |
90 |
0.49 |
Binding ≤ 10μM
|
|
BLKPC-1-B |
Carbepenem-hydrolyzing Beta-lactamase KPC (cluster #1 Of 1), Bacterial |
Bacteria |
374 |
0.45 |
Binding ≤ 10μM
|
|
D1MIX9-1-B |
Beta-lactamase GES-13 (cluster #1 Of 1), Bacterial |
Bacteria |
60 |
0.51 |
Binding ≤ 10μM
|
|
Q2XPY6-1-B |
Extended-spectrum Beta-lactamase CTX-M-53 (cluster #1 Of 1), Bacterial |
Bacteria |
2 |
0.61 |
Binding ≤ 10μM
|
|
Q46991-1-B |
IMI-1 (cluster #1 Of 1), Bacterial |
Bacteria |
30 |
0.53 |
Binding ≤ 10μM
|
|
Q50H31-1-B |
Class D Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
160 |
0.48 |
Binding ≤ 10μM
|
|
Q59401-1-B |
Beta-lactamase Class C (cluster #1 Of 1), Bacterial |
Bacteria |
3400 |
0.38 |
Binding ≤ 10μM
|
|
Q6GWS8-1-B |
Beta-lactamase TEM-125 (cluster #1 Of 1), Bacterial |
Bacteria |
240 |
0.46 |
Binding ≤ 10μM
|
|
Q6JP75-1-B |
Carbapenem-hydrolizing Beta-lactamase SFC-1 (cluster #1 Of 1), Bacterial |
Bacteria |
6900 |
0.36 |
Binding ≤ 10μM
|
|
Q6W9J1-1-B |
Beta-lactamase TEM (cluster #1 Of 1), Bacterial |
Bacteria |
60 |
0.51 |
Binding ≤ 10μM
|
|
Q6W9J1-1-B |
Beta-lactamase TEM (cluster #1 Of 1), Bacterial |
Bacteria |
320 |
0.45 |
Binding ≤ 10μM
|
|
Q9EXV5-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
2 |
0.61 |
Binding ≤ 10μM
|
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
830 |
0.43 |
Functional ≤ 10μM
|
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
943 |
0.42 |
Functional ≤ 10μM
|
|
BLAT-1-B |
Beta-lactamase TEM (cluster #1 Of 1), Bacterial |
Bacteria |
25 |
0.53 |
Functional ≤ 10μM
|
|
BLO1-1-B |
Beta-lactamase OXA-1 (cluster #1 Of 1), Bacterial |
Bacteria |
4800 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.22 |
-0.82 |
-55.23 |
0 |
9 |
-1 |
125 |
299.288 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-5-B |
Beta-lactamase (cluster #5 Of 6), Bacterial |
Bacteria |
5000 |
0.22 |
Binding ≤ 10μM
|
|
AMPH-1-B |
Penicillin-binding Protein AmpH (cluster #1 Of 2), Bacterial |
Bacteria |
5000 |
0.22 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
38 |
0.31 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.26 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.26 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
33 |
0.32 |
Binding ≤ 10μM
|
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KS6A3-1-E |
Ribosomal Protein S6 Kinase Alpha 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
KS6A5-1-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
|
KS6B1-1-E |
Ribosomal Protein S6 Kinase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.33 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
|
SIRT1-1-E |
NAD-dependent Deacetylase Sirtuin 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5100 |
0.22 |
Binding ≤ 10μM
|
|
SIRT2-3-E |
NAD-dependent Deacetylase Sirtuin 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.26 |
Binding ≤ 10μM |
|
SIRT3-2-E |
NAD-dependent Deacetylase Sirtuin 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.23 |
Binding ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1960 |
0.24 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
840 |
0.26 |
Functional ≤ 10μM
|
|
Z81252-1-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #1 Of 11), Other |
Other |
1770 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
10.08 |
-46.88 |
5 |
7 |
1 |
111 |
458.567 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.47 |
10.2 |
-15.14 |
4 |
7 |
0 |
110 |
457.559 |
7 |
↓
|
|
|
|
Analogs
-
4554508
-
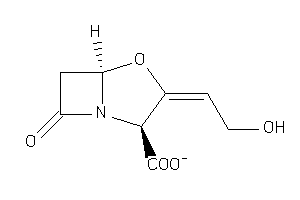
-
4554509
-
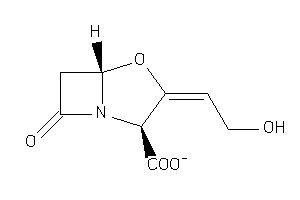
-
4554510
-
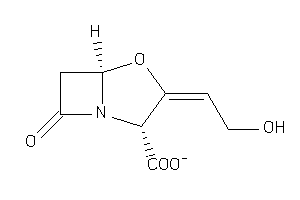
-
4554511
-
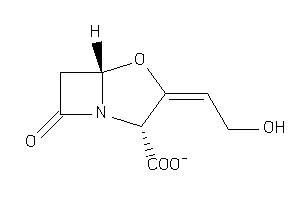
-
1169
-
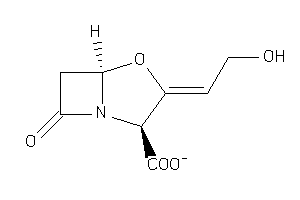
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A0ZX81-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
1720 |
0.58 |
Binding ≤ 10μM
|
|
A1E3K9-1-B |
Beta-lactamase SCO-1 (cluster #1 Of 1), Bacterial |
Bacteria |
70 |
0.72 |
Binding ≤ 10μM
|
|
A4KCT8-1-B |
Gil1 (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.80 |
Binding ≤ 10μM
|
|
A8RR46-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
85 |
0.71 |
Binding ≤ 10μM
|
|
AMPC-5-B |
Beta-lactamase (cluster #5 Of 6), Bacterial |
Bacteria |
800 |
0.61 |
Binding ≤ 10μM
|
|
BLA1-1-B |
Beta-lactamase L1 (cluster #1 Of 3), Bacterial |
Bacteria |
170 |
0.68 |
Binding ≤ 10μM
|
|
BLAC-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
80 |
0.71 |
Binding ≤ 10μM
|
|
BLAT-3-B |
Beta-lactamase TEM (cluster #3 Of 4), Bacterial |
Bacteria |
800 |
0.61 |
Binding ≤ 10μM
|
|
BLO1-1-B |
Beta-lactamase OXA-1 (cluster #1 Of 1), Bacterial |
Bacteria |
3200 |
0.55 |
Binding ≤ 10μM
|
|
D1MIX9-1-B |
Beta-lactamase GES-13 (cluster #1 Of 1), Bacterial |
Bacteria |
100 |
0.70 |
Binding ≤ 10μM
|
|
Q2XPY6-1-B |
Extended-spectrum Beta-lactamase CTX-M-53 (cluster #1 Of 1), Bacterial |
Bacteria |
10 |
0.80 |
Binding ≤ 10μM
|
|
Q4TVR4-1-B |
Beta-lactamase SHV-55 (cluster #1 Of 1), Bacterial |
Bacteria |
20 |
0.77 |
Binding ≤ 10μM
|
|
Q50H31-1-B |
Class D Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
2000 |
0.57 |
Binding ≤ 10μM
|
|
Q6GWS8-1-B |
Beta-lactamase TEM-125 (cluster #1 Of 1), Bacterial |
Bacteria |
8600 |
0.51 |
Binding ≤ 10μM
|
|
Q9EXV5-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.80 |
Binding ≤ 10μM
|
|
BLAT-1-B |
Beta-lactamase TEM (cluster #1 Of 1), Bacterial |
Bacteria |
60 |
0.72 |
Functional ≤ 10μM
|
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
40 |
0.74 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.42 |
-3.38 |
-49.41 |
1 |
6 |
-1 |
90 |
198.154 |
2 |
↓
|
|
|
|
Analogs
-
17194628
-
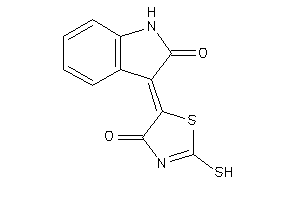
-
33929197
-
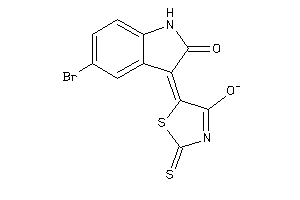
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-5-B |
Beta-lactamase (cluster #5 Of 6), Bacterial |
Bacteria |
2600 |
0.46 |
Binding ≤ 10μM
|
|
Q6W9J1-1-B |
Beta-lactamase TEM (cluster #1 Of 1), Bacterial |
Bacteria |
8700 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
2.85 |
-47.62 |
1 |
4 |
-1 |
69 |
261.307 |
0 |
↓
|
|
|
|
Analogs
-
2086
-
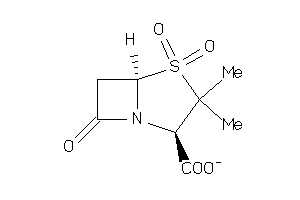
Draw
Identity
99%
90%
80%
70%
Vendors
And 58 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A1E3K9-1-B |
Beta-lactamase SCO-1 (cluster #1 Of 1), Bacterial |
Bacteria |
6200 |
0.49 |
Binding ≤ 10μM
|
|
A8RR46-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
75 |
0.66 |
Binding ≤ 10μM
|
|
AMPC-5-B |
Beta-lactamase (cluster #5 Of 6), Bacterial |
Bacteria |
1400 |
0.55 |
Binding ≤ 10μM
|
|
BLAC-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
6500 |
0.48 |
Binding ≤ 10μM
|
|
BLAT-3-B |
Beta-lactamase TEM (cluster #3 Of 4), Bacterial |
Bacteria |
800 |
0.57 |
Binding ≤ 10μM
|
|
D1MIX9-1-B |
Beta-lactamase GES-13 (cluster #1 Of 1), Bacterial |
Bacteria |
370 |
0.60 |
Binding ≤ 10μM
|
|
D5HSX5-1-B |
Extended-spectrum Beta-lactamase PER-6 (cluster #1 Of 1), Bacterial |
Bacteria |
4000 |
0.50 |
Binding ≤ 10μM
|
|
Q0VTR6-1-B |
ADC-14 Protein (cluster #1 Of 1), Bacterial |
Bacteria |
1120 |
0.56 |
Binding ≤ 10μM
|
|
Q0VTR8-1-B |
ADC-16 Protein (cluster #1 Of 1), Bacterial |
Bacteria |
7758 |
0.48 |
Binding ≤ 10μM
|
|
Q7X575-1-B |
Beta-lactamase SHV-11 (cluster #1 Of 1), Bacterial |
Bacteria |
2400 |
0.52 |
Binding ≤ 10μM
|
|
Z50185-1-O |
Staphylococcus Aureus (cluster #1 Of 1), Other |
Other |
6800 |
0.48 |
Binding ≤ 10μM |
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 1), Other |
Other |
1900 |
0.53 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-0.6 |
-51.76 |
0 |
6 |
-1 |
95 |
232.237 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.65 |
6.84 |
-38.29 |
3 |
3 |
1 |
46 |
292.443 |
7 |
↓
|
|
|
|
Analogs
-
1227821
-
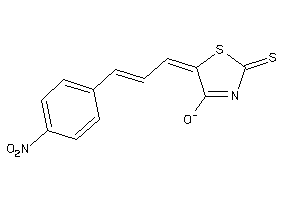
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-5-B |
Beta-lactamase (cluster #5 Of 6), Bacterial |
Bacteria |
4200 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
6.4 |
-43.77 |
0 |
5 |
-1 |
82 |
291.333 |
3 |
↓
|
|
|
|
Analogs
-
33247330
-
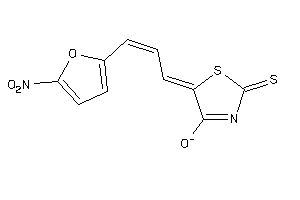
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-5-B |
Beta-lactamase (cluster #5 Of 6), Bacterial |
Bacteria |
450 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
2.94 |
-54.81 |
0 |
6 |
-1 |
95 |
281.294 |
3 |
↓
|
|