|
|
|
|
|
Analogs
-
4019237
-
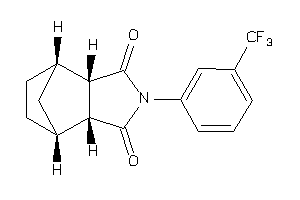
-
4019238
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
360 |
0.41 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Functional ≤ 10μM
|
|
Z80243-1-O |
MDA-MB-453 (Breast Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
317 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
8.77 |
-10.1 |
0 |
3 |
0 |
37 |
309.287 |
2 |
↓
|
|
|
|
Analogs
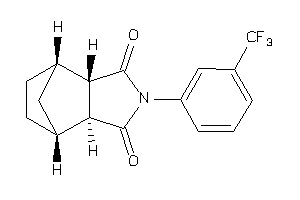
-
4019238
-

-
2068322
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
360 |
0.41 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Functional ≤ 10μM
|
|
Z80243-1-O |
MDA-MB-453 (Breast Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
317 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
8.52 |
-10.63 |
0 |
3 |
0 |
37 |
309.287 |
2 |
↓
|
|
|
|
Analogs
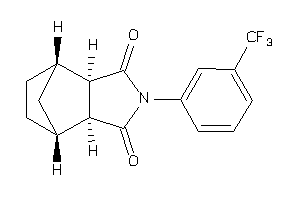
-
897408
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 40 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
11 |
0.38 |
Binding ≤ 10μM
|
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
670 |
0.30 |
Binding ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7200 |
0.25 |
Binding ≤ 10μM
|
|
Q5EM59-1-E |
Androgen Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.29 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.32 |
Functional ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
890 |
0.29 |
Functional ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1819 |
0.28 |
Functional ≤ 10μM
|
|
Z80243-1-O |
MDA-MB-453 (Breast Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
173 |
0.33 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
870 |
0.29 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
1000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
4.05 |
-16.95 |
2 |
6 |
0 |
107 |
430.379 |
6 |
↓
|
|
|
|
Analogs
-
538564
-
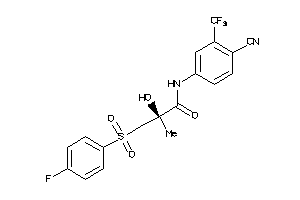
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
4.04 |
-16.77 |
2 |
6 |
0 |
107 |
430.379 |
6 |
↓
|
|
|
|
Analogs
-
36805638
-
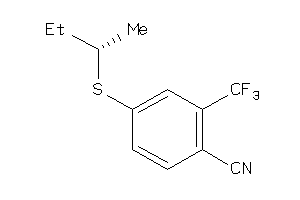
-
34960782
-
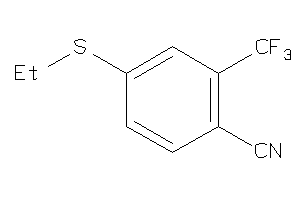
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
315 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
8.35 |
-6.26 |
0 |
1 |
0 |
24 |
259.296 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
225 |
0.47 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
123 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
10.35 |
-6.95 |
0 |
1 |
0 |
24 |
293.313 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
56 |
0.48 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
9.12 |
-8.45 |
0 |
2 |
0 |
33 |
309.312 |
4 |
↓
|
|
|
|
Analogs
-
36805638
-

-
34960782
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
23 |
0.67 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.70 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
8.33 |
-6.66 |
0 |
1 |
0 |
24 |
245.269 |
4 |
↓
|
|
|
|
Analogs
-
36805638
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
49 |
0.60 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
838 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.39 |
9.11 |
-6.72 |
0 |
1 |
0 |
24 |
259.296 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
83 |
0.47 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
8.48 |
-8.99 |
0 |
3 |
0 |
42 |
293.244 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
36805620
-
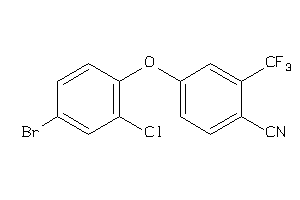
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2720 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.92 |
9.11 |
-7.35 |
0 |
2 |
0 |
33 |
297.663 |
3 |
↓
|
|
|
|
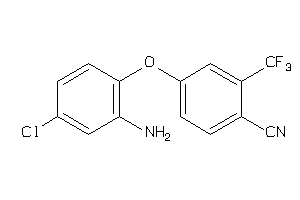
Analogs
-
36667217
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6650 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.95 |
9.12 |
-7.27 |
0 |
2 |
0 |
33 |
297.663 |
3 |
↓
|
|
|
|
|
|
|
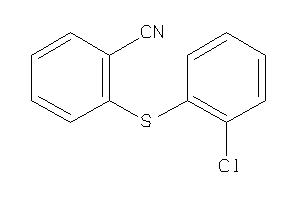
Analogs
-
21798825
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.61 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
78 |
0.59 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
9.99 |
-5.77 |
0 |
1 |
0 |
24 |
259.761 |
2 |
↓
|
|
|
|
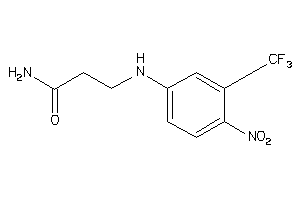
Analogs
-
36747497
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
3.1 |
-9.74 |
2 |
6 |
0 |
95 |
292.213 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.39 |
9.26 |
-58.21 |
6 |
8 |
1 |
129 |
493.591 |
6 |
↓
|
|
|
|
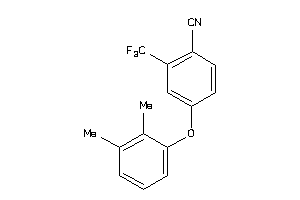
Analogs
-
36805614
-
-
21819829
-
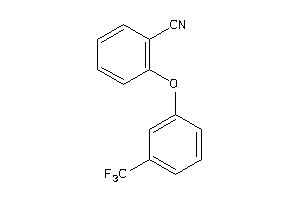
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
92 |
0.49 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
743 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
9.3 |
-6.99 |
0 |
2 |
0 |
33 |
277.245 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.56 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
90 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
6.63 |
-6.62 |
1 |
2 |
0 |
44 |
269.266 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
74 |
0.48 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
22 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.66 |
9.63 |
-8.57 |
0 |
2 |
0 |
33 |
309.312 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
184 |
0.47 |
Binding ≤ 10μM
|
|
PRGR-3-E |
Progesterone Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6950 |
0.36 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
54 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.60 |
9.82 |
-8.22 |
0 |
1 |
0 |
24 |
297.276 |
3 |
↓
|
|
|
|
Analogs
-
36669161
-
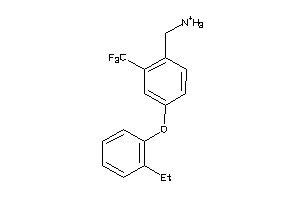
-
21819829
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
20 |
0.51 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
356 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
10.06 |
-6.85 |
0 |
2 |
0 |
33 |
291.272 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.41 |
Binding ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
2.45 |
-11.43 |
1 |
6 |
0 |
84 |
369.343 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.96 |
1.44 |
-16.2 |
1 |
5 |
0 |
67 |
385.411 |
6 |
↓
|
|
|
|
Analogs
-
36886878
-
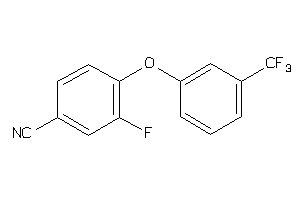
-
36749907
-
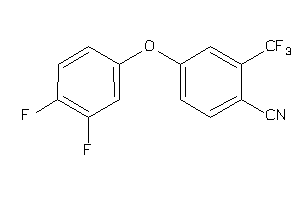
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
488 |
0.42 |
Binding ≤ 10μM
|
|
ANDR-3-E |
Androgen Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
8.65 |
-8.28 |
0 |
2 |
0 |
33 |
299.198 |
3 |
↓
|
|
|
|
Analogs
-
33865679
-
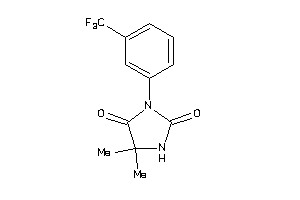
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-4-E |
Androgen Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
5.04 |
-9.41 |
1 |
5 |
0 |
73 |
297.236 |
2 |
↓
|
|