|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
3220 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
7.66 |
-10.17 |
0 |
4 |
0 |
52 |
288.306 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
920 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
2.68 |
-9.06 |
0 |
3 |
0 |
42 |
294.26 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.35 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
1.56 |
-12.61 |
0 |
5 |
0 |
61 |
318.332 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
920 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
1.06 |
-8.83 |
0 |
3 |
0 |
42 |
292.725 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.40 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
660 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.47 |
0.45 |
-10.24 |
0 |
3 |
0 |
42 |
337.176 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
9900 |
0.33 |
Binding ≤ 10μM
|
|
HPGDS-1-E |
Hematopoietic Prostaglandin D Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3800 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
5.51 |
-10.78 |
1 |
4 |
0 |
63 |
274.279 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
280 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
3.38 |
-11.25 |
0 |
3 |
0 |
42 |
326.277 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.96 |
2.54 |
-7.62 |
2 |
3 |
0 |
37 |
202.257 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
-2.75 |
-6.08 |
1 |
2 |
0 |
28 |
170.215 |
0 |
↓
|
|
|
|
Analogs
-
32009926
-
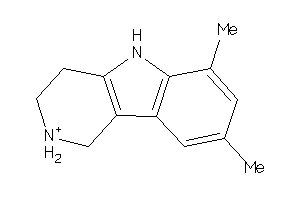
-
34602288
-
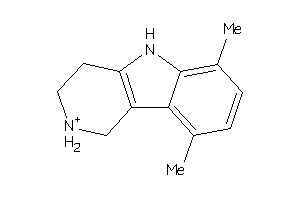
-
34985412
-
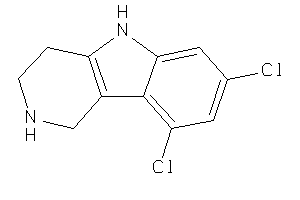
Draw
Identity
99%
90%
80%
70%
Vendors
And 64 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.93 |
3.2 |
-4.94 |
2 |
2 |
0 |
28 |
172.231 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.93 |
3.26 |
-5.76 |
2 |
2 |
0 |
28 |
172.231 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
84 |
0.76 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
-0.23 |
-47.82 |
4 |
4 |
1 |
70 |
200.649 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.39 |
-0.61 |
-5.96 |
3 |
4 |
0 |
68 |
199.641 |
3 |
↓
|
|
|
|
Analogs
-
4713053
-
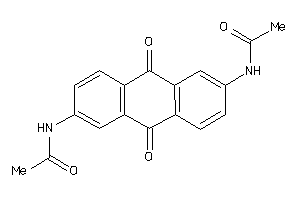
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
6.24 |
-15.88 |
1 |
4 |
0 |
63 |
265.268 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.40 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1330 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
0.45 |
-8.78 |
0 |
3 |
0 |
42 |
337.176 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
900 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
1.06 |
-10.23 |
0 |
3 |
0 |
42 |
292.725 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.44 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
0.36 |
-9.34 |
0 |
3 |
0 |
42 |
264.309 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
920 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
8.42 |
-9.2 |
0 |
3 |
0 |
43 |
276.27 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6500 |
0.45 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
3600 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
5.82 |
-9.7 |
0 |
5 |
0 |
67 |
233.662 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.66 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.66 |
Binding ≤ 10μM
|
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1290 |
0.59 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1290 |
0.59 |
Binding ≤ 10μM
|
|
NISCH-2-E |
Nischarin (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1290 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
3.73 |
-5.25 |
2 |
2 |
0 |
28 |
251.127 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
240 |
0.62 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
3830 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
5.87 |
-6.42 |
1 |
2 |
0 |
17 |
198.269 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
9400 |
0.37 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6500 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
1.55 |
-15.83 |
1 |
5 |
0 |
54 |
283.759 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.96 |
3.87 |
-48.25 |
3 |
3 |
1 |
42 |
203.265 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
5.5 |
-48.98 |
2 |
2 |
1 |
22 |
173.239 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.63 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
210 |
0.67 |
Binding ≤ 10μM
|
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
2785 |
0.56 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
2785 |
0.56 |
Binding ≤ 10μM
|
|
NISCH-2-E |
Nischarin (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2785 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
3.77 |
-7.1 |
2 |
2 |
0 |
28 |
251.127 |
0 |
↓
|
|
|
|
Analogs
-
19855507
-
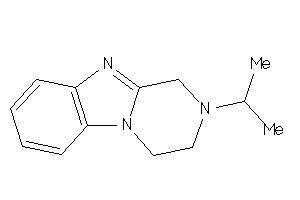
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6790 |
0.56 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6790 |
0.56 |
Binding ≤ 10μM
|
|
NISCH-3-E |
Nischarin (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6790 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.02 |
4.42 |
-53.02 |
2 |
3 |
1 |
34 |
174.227 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
6.62 |
-46.29 |
2 |
2 |
1 |
22 |
201.293 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
-3.5 |
-5.81 |
1 |
2 |
0 |
28 |
249.111 |
0 |
↓
|
|
|
|
Analogs
-
12966
-
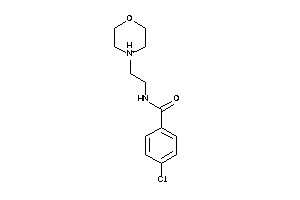
-
6092705
-
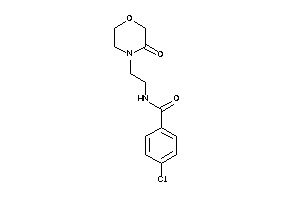
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5 |
0.65 |
Binding ≤ 10μM |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1080 |
0.46 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
2.6 |
-8.83 |
1 |
4 |
0 |
42 |
268.744 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 53 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
832 |
0.34 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Binding ≤ 10μM
|
|
PPARD-2-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3620 |
0.30 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.44 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.40 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
325 |
0.36 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.40 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
210 |
0.37 |
Functional ≤ 10μM
|
|
Z80106-1-O |
COS-1 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.43 |
Functional ≤ 10μM
|
|
Z80169-1-O |
Huh-7 (Hepatocellular Carcinoma) (cluster #1 Of 1), Other |
Other |
220 |
0.37 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z81117-1-O |
Keratinocytes (Keratinocytes) (cluster #1 Of 2), Other |
Other |
8000 |
0.29 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
5000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
7.4 |
-33.54 |
2 |
6 |
1 |
73 |
358.443 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.35 |
7.28 |
-11.07 |
1 |
6 |
0 |
72 |
357.435 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 54 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
832 |
0.34 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Binding ≤ 10μM
|
|
PPARD-2-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3620 |
0.30 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.44 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.41 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
325 |
0.36 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.40 |
Functional ≤ 10μM
|
|
Z80106-1-O |
COS-1 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.43 |
Functional ≤ 10μM
|
|
Z80169-1-O |
Huh-7 (Hepatocellular Carcinoma) (cluster #1 Of 1), Other |
Other |
220 |
0.37 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z81117-1-O |
Keratinocytes (Keratinocytes) (cluster #1 Of 2), Other |
Other |
8000 |
0.29 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
5000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
7.4 |
-33.23 |
2 |
6 |
1 |
73 |
358.443 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.35 |
7.28 |
-11.54 |
1 |
6 |
0 |
72 |
357.435 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6880 |
0.36 |
Binding ≤ 10μM |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
195 |
0.47 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.46 |
3.88 |
-46.57 |
3 |
5 |
1 |
70 |
292.384 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.46 |
3.5 |
-8.8 |
2 |
5 |
0 |
68 |
291.376 |
4 |
↓
|
|
|
|
Analogs
-
40072708
-
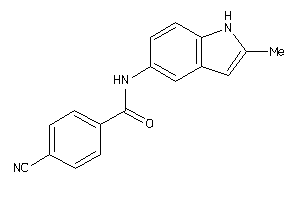
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
3840 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
7.11 |
-13.5 |
2 |
3 |
0 |
45 |
250.301 |
2 |
↓
|
|
|
|
Analogs
-
32818533
-
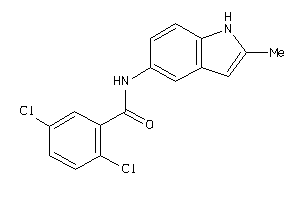
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8180 |
0.36 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
760 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
7.62 |
-14.33 |
2 |
3 |
0 |
45 |
284.746 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
2911 |
0.39 |
Binding ≤ 10μM |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
214 |
0.47 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
9.83 |
-6.83 |
1 |
3 |
0 |
37 |
305.834 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1752 |
0.40 |
Binding ≤ 10μM |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
9.38 |
-7.23 |
1 |
3 |
0 |
37 |
289.379 |
3 |
↓
|
|
|
|
Analogs
-
35469789
-

-
35469790
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
3689 |
0.38 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.73 |
10.15 |
-8.44 |
1 |
3 |
0 |
41 |
285.416 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.80 |
10.33 |
-24.22 |
2 |
3 |
1 |
39 |
286.424 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
363 |
0.39 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
10.55 |
-9.84 |
1 |
6 |
0 |
83 |
330.413 |
4 |
↓
|
|
|
|
Analogs
-
192778
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
11 |
0.48 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.82 |
-0.15 |
-9.09 |
1 |
6 |
0 |
83 |
330.413 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
204 |
0.45 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.64 |
10.77 |
-7.45 |
1 |
3 |
0 |
37 |
340.279 |
3 |
↓
|
|
|
|
Analogs
-
40148061
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.29 |
9.95 |
-7.54 |
1 |
3 |
0 |
37 |
303.406 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.29 |
10.28 |
-30.79 |
2 |
3 |
1 |
39 |
304.414 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8128 |
0.34 |
Binding ≤ 10μM |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5495 |
0.35 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.50 |
10.8 |
-9.01 |
1 |
3 |
0 |
41 |
299.443 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.50 |
10.85 |
-26.87 |
2 |
3 |
1 |
42 |
300.451 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5570 |
0.32 |
Binding ≤ 10μM |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
2943 |
0.34 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.24 |
6.85 |
-8.29 |
0 |
7 |
0 |
71 |
314.345 |
4 |
↓
|
|
|
|
Analogs
-
36667185
-

-
36984666
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
7840 |
0.34 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
5.69 |
-8.33 |
0 |
7 |
0 |
71 |
286.291 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4620 |
0.39 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
44 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
5.23 |
-13.55 |
1 |
4 |
0 |
59 |
253.257 |
3 |
↓
|
|
|
|
Analogs
-
32625194
-
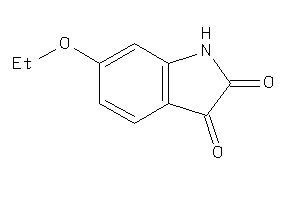
-
5170575
-
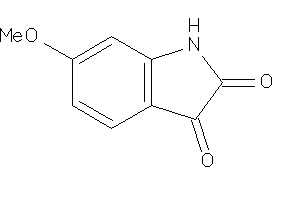
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
59 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
5.27 |
-11.16 |
1 |
4 |
0 |
59 |
253.257 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
9440 |
0.39 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
663 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
4.8 |
-11.49 |
1 |
4 |
0 |
59 |
239.23 |
2 |
↓
|
|
|
|
Analogs
-
16946225
-

-
16946229
-

-
16946294
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
562 |
0.51 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
513 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
5.39 |
-10.36 |
1 |
3 |
0 |
50 |
223.231 |
1 |
↓
|
|