|
|
Analogs
-
4026300
-
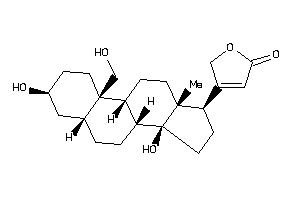
-
4026301
-
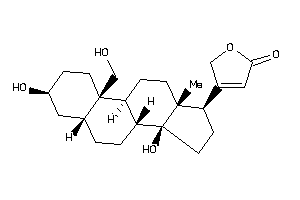
-
4073972
-
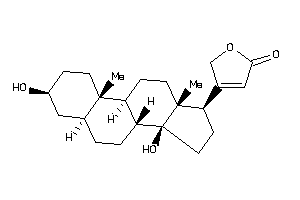
-
4073973
-
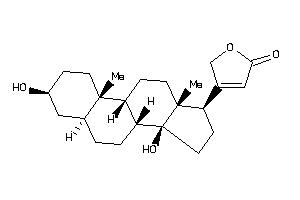
-
5696447
-
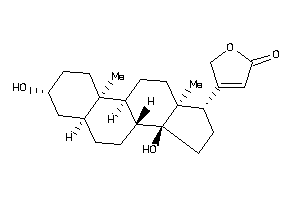
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT12A-1-E |
Potassium-transporting ATPase Alpha Chain 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.37 |
Binding ≤ 10μM
|
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
63 |
0.37 |
Binding ≤ 10μM |
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
63 |
0.37 |
Binding ≤ 10μM |
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
Z100253-1-O |
Monoclonal Antibody (mAb) (cluster #1 Of 1), Other |
Other |
880 |
0.31 |
Binding ≤ 10μM
|
|
Z50512-4-O |
Cavia Porcellus (cluster #4 Of 7), Other |
Other |
600 |
0.32 |
Functional ≤ 10μM
|
|
Z80164-2-O |
HT-1080 (Fibrosarcoma Cells) (cluster #2 Of 6), Other |
Other |
1500 |
0.30 |
Functional ≤ 10μM
|
|
Z80224-2-O |
MCF7 (Breast Carcinoma Cells) (cluster #2 Of 14), Other |
Other |
600 |
0.32 |
Functional ≤ 10μM
|
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
620 |
0.32 |
Functional ≤ 10μM
|
|
Z81115-4-O |
KB (Squamous Cell Carcinoma) (cluster #4 Of 6), Other |
Other |
9610 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
6.57 |
-10.93 |
2 |
4 |
0 |
67 |
374.521 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
-1.85 |
-11.16 |
2 |
4 |
0 |
66 |
374.521 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
-1.3 |
-10.49 |
2 |
4 |
0 |
66 |
374.521 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT12A-1-E |
Potassium-transporting ATPase Alpha Chain 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.30 |
Binding ≤ 10μM
|
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.28 |
-3.02 |
-5.88 |
3 |
4 |
0 |
73 |
374.521 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6310 |
0.28 |
Binding ≤ 10μM |
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6310 |
0.28 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
9.02 |
-11.42 |
1 |
3 |
0 |
47 |
358.522 |
1 |
↓
|
|