|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B2CL1-1-E |
Apoptosis Regulator Bcl-X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
630 |
0.24 |
Binding ≤ 10μM
|
|
B2LA1-1-E |
Bcl-2-related Protein A1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2170 |
0.22 |
Binding ≤ 10μM
|
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.25 |
Binding ≤ 10μM
|
|
MCL1-1-E |
Induced Myeloid Leukemia Cell Differentiation Protein Mcl-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
540 |
0.24 |
Binding ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
1460 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.21 |
5.79 |
-51.8 |
3 |
8 |
-1 |
152 |
489.5 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.21 |
6.59 |
-87.22 |
2 |
8 |
-2 |
155 |
488.492 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.21 |
5.67 |
-44.87 |
3 |
8 |
-1 |
152 |
489.5 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B2CL1-1-E |
Apoptosis Regulator Bcl-X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6580 |
0.18 |
Binding ≤ 10μM
|
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.21 |
Binding ≤ 10μM
|
|
MCL1-1-E |
Induced Myeloid Leukemia Cell Differentiation Protein Mcl-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
260 |
0.22 |
Binding ≤ 10μM
|
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.23 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
200 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.96 |
10.32 |
-26.18 |
4 |
7 |
0 |
124 |
573.711 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
7.96 |
11.28 |
-68.52 |
3 |
7 |
-1 |
127 |
572.703 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1288 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.68 |
-1.19 |
-22.6 |
2 |
9 |
0 |
123 |
511.581 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1288 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.68 |
9.77 |
-17.14 |
2 |
9 |
0 |
124 |
511.581 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BAD-1-E |
Bcl2-antagonist Of Cell Death (BAD) (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1110 |
0.24 |
Binding ≤ 10μM
|
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
170 |
0.27 |
Binding ≤ 10μM
|
|
MCL1-1-E |
Induced Myeloid Leukemia Cell Differentiation Protein Mcl-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.30 |
Binding ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
260 |
0.26 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
120 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.89 |
9.85 |
-135.55 |
1 |
7 |
-2 |
123 |
481.485 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.89 |
8.88 |
-48.75 |
2 |
7 |
-1 |
120 |
482.493 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.89 |
9.68 |
-54.61 |
2 |
7 |
-1 |
120 |
482.493 |
6 |
↓
|
|
|
|
Analogs
-
3978503
-
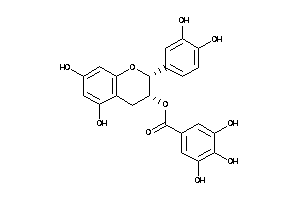
-
4534390
-
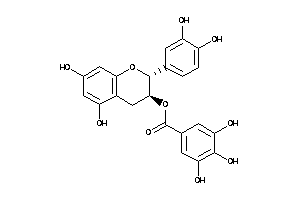
-
4544252
-
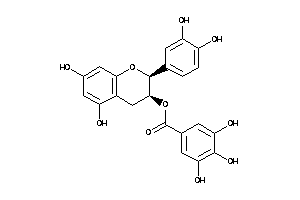
-
8681494
-
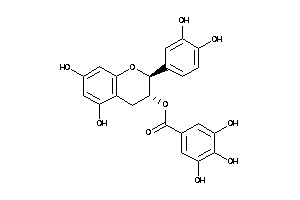
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
6PGD-1-E |
6-phosphogluconate Dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
720 |
0.26 |
Binding ≤ 10μM
|
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
234 |
0.28 |
Binding ≤ 10μM
|
|
ERG1-3-E |
Squalene Monooxygenase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
690 |
0.26 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6026 |
0.22 |
Binding ≤ 10μM
|
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.23 |
Binding ≤ 10μM
|
|
MK14-2-E |
MAP Kinase P38 Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2210 |
0.24 |
Binding ≤ 10μM
|
|
MMP14-1-E |
Matrix Metalloproteinase 14 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6800 |
0.22 |
Binding ≤ 10μM
|
|
MMP2-2-E |
72 KDa Type IV Collagenase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9600 |
0.21 |
Binding ≤ 10μM
|
|
Q965D5-1-E |
Enoyl-acyl-carrier Protein Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
|
Q965D6-1-E |
3-oxoacyl-acyl-carrier Protein Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.28 |
Binding ≤ 10μM
|
|
Q965D7-2-E |
Fatty Acid Synthase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.27 |
Binding ≤ 10μM
|
|
G6PD-1-F |
Glucose-6-phosphate 1-dehydrogenase (cluster #1 Of 1), Fungal |
Fungi |
250 |
0.28 |
Binding ≤ 10μM
|
|
Z50142-1-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #1 Of 2), Other |
Other |
391 |
0.27 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
9900 |
0.21 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
9400 |
0.21 |
Functional ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
730 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
-6.71 |
-20.66 |
8 |
11 |
0 |
197 |
458.375 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.25 |
-5.81 |
-60.24 |
7 |
11 |
-1 |
200 |
457.367 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.25 |
-5.67 |
-56.02 |
7 |
11 |
-1 |
200 |
457.367 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
5835087
-
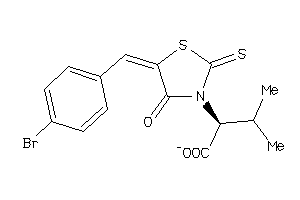
-
5835550
-
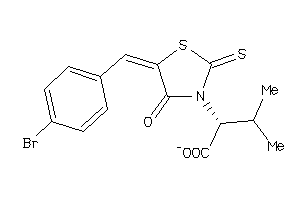
-
1227906
-
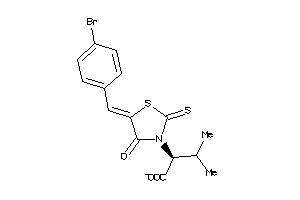
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B2CL1-1-E |
Apoptosis Regulator Bcl-X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.32 |
Binding ≤ 10μM
|
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
11.36 |
-52.69 |
0 |
4 |
-1 |
62 |
399.311 |
4 |
↓
|
|
|
|
Analogs
-
4534390
-

-
4544252
-

-
8681494
-

-
3870415
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
6PGD-1-E |
6-phosphogluconate Dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
210 |
0.29 |
Binding ≤ 10μM
|
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
229 |
0.29 |
Binding ≤ 10μM
|
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.22 |
Binding ≤ 10μM
|
|
MK14-2-E |
MAP Kinase P38 Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1470 |
0.26 |
Binding ≤ 10μM
|
|
Q965D5-1-E |
Enoyl-acyl-carrier Protein Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.28 |
Binding ≤ 10μM
|
|
Q965D6-1-E |
3-oxoacyl-acyl-carrier Protein Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.26 |
Binding ≤ 10μM
|
|
Q965D7-2-E |
Fatty Acid Synthase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.28 |
Binding ≤ 10μM
|
|
FABI-1-B |
Enoyl-[acyl-carrier-protein] Reductase (cluster #1 Of 2), Bacterial |
Bacteria |
10000 |
0.22 |
Binding ≤ 10μM
|
|
G6PD-1-F |
Glucose-6-phosphate 1-dehydrogenase (cluster #1 Of 1), Fungal |
Fungi |
180 |
0.30 |
Binding ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
760 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
-4.55 |
-20.07 |
7 |
10 |
0 |
177 |
442.376 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.54 |
-3.63 |
-57.28 |
6 |
10 |
-1 |
180 |
441.368 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.54 |
-3.53 |
-54.34 |
6 |
10 |
-1 |
180 |
441.368 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B2CL1-1-E |
Apoptosis Regulator Bcl-X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
652 |
0.22 |
Binding ≤ 10μM
|
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
192 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.57 |
10.53 |
-58.63 |
1 |
9 |
-1 |
131 |
567.713 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.57 |
10.78 |
-48.83 |
3 |
9 |
1 |
129 |
569.729 |
10 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 28 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALDR-4-E |
Aldose Reductase (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
500 |
0.23 |
Binding ≤ 10μM
|
|
B2CL1-1-E |
Apoptosis Regulator Bcl-X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
540 |
0.23 |
Binding ≤ 10μM
|
|
BAD-1-E |
Bcl2-antagonist Of Cell Death (BAD) (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1710 |
0.21 |
Binding ≤ 10μM
|
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.23 |
Binding ≤ 10μM
|
|
MCL1-1-E |
Induced Myeloid Leukemia Cell Differentiation Protein Mcl-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6543 |
0.19 |
Binding ≤ 10μM
|
|
MDHC-1-E |
Malate Dehydrogenase Cytoplasmic (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5800 |
0.19 |
Binding ≤ 10μM
|
|
MDHM-2-E |
Malate Dehydrogenase, Mitochondrial (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.20 |
Binding ≤ 10μM
|
|
Q0PJ46-1-E |
Lactate Dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2640 |
0.21 |
Binding ≤ 10μM
|
|
Q6VVP7-1-E |
Malate Dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2030 |
0.21 |
Binding ≤ 10μM
|
|
Z100733-1-O |
CT26 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2210 |
0.21 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7000 |
0.19 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
9140 |
0.19 |
Functional ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
8400 |
0.19 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
1320 |
0.22 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
3330 |
0.20 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
3700 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.57 |
5.14 |
-19.48 |
6 |
8 |
0 |
156 |
518.562 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.57 |
6.21 |
-32.82 |
5 |
8 |
0 |
158 |
517.554 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5800 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
-4.36 |
-41.01 |
6 |
6 |
1 |
112 |
330.371 |
3 |
↓
|
|