|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
173 |
1.05 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
148 |
1.06 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.30 |
-2.79 |
-9.58 |
3 |
4 |
0 |
72 |
152.219 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
145 |
0.56 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
8 |
0.67 |
Binding ≤ 10μM
|
|
CAH9-6-E |
Carbonic Anhydrase IX (cluster #6 Of 11), Eukaryotic |
Eukaryotes |
10 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.14 |
-12.84 |
-19.91 |
4 |
8 |
0 |
138 |
290.363 |
10 |
↓
|
|
|
|
Analogs
-
21984495
-
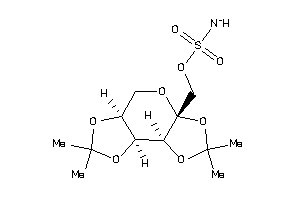
-
22061272
-
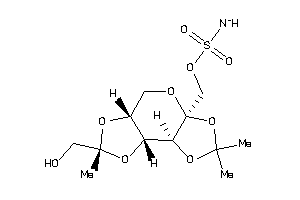
-
22061274
-
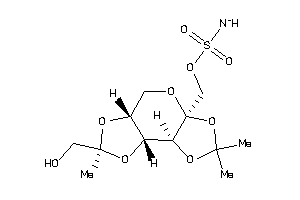
-
23586802
-
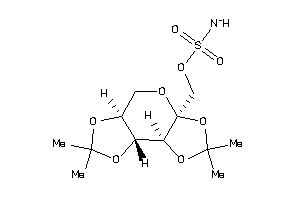
-
23586806
-
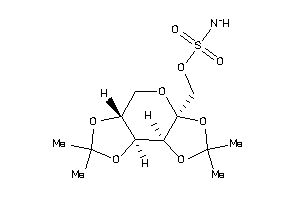
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-2-A |
Carbonic Anhydrase (cluster #2 Of 2), Archaea |
Archaea |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CYNT-3-B |
Carbonic Anhydrase (cluster #3 Of 3), Bacterial |
Bacteria |
474 |
0.40 |
Binding ≤ 10μM
|
|
P96878-2-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #2 Of 2), Bacterial |
Bacteria |
3020 |
0.35 |
Binding ≤ 10μM
|
|
Y1284-2-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #2 Of 2), Bacterial |
Bacteria |
612 |
0.40 |
Binding ≤ 10μM
|
|
B5SU02-4-E |
Alpha Carbonic Anhydrase (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
29 |
0.48 |
Binding ≤ 10μM
|
|
C0IX24-3-E |
Carbonic Anhydrase (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
367 |
0.41 |
Binding ≤ 10μM
|
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CAH12-6-E |
Carbonic Anhydrase XII (cluster #6 Of 9), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
CAH13-3-E |
Carbonic Anhydrase XIII (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
47 |
0.47 |
Binding ≤ 10μM
|
|
CAH14-5-E |
Carbonic Anhydrase XIV (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1460 |
0.37 |
Binding ≤ 10μM
|
|
CAH15-5-E |
Carbonic Anhydrase 15 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CAH3-4-E |
Carbonic Anhydrase III (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH4-7-E |
Carbonic Anhydrase IV (cluster #7 Of 16), Eukaryotic |
Eukaryotes |
54 |
0.46 |
Binding ≤ 10μM
|
|
CAH5A-3-E |
Carbonic Anhydrase VA (cluster #3 Of 10), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH5B-3-E |
Carbonic Anhydrase VB (cluster #3 Of 9), Eukaryotic |
Eukaryotes |
30 |
0.48 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH7-5-E |
Carbonic Anhydrase VII (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
CAH9-7-E |
Carbonic Anhydrase IX (cluster #7 Of 11), Eukaryotic |
Eukaryotes |
58 |
0.46 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.42 |
Functional ≤ 10μM
|
|
CAH2-2-E |
Carbonic Anhydrase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Functional ≤ 10μM
|
|
CAH4-2-E |
Carbonic Anhydrase IV (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
54 |
0.46 |
Functional ≤ 10μM
|
|
CAN-2-F |
Carbonic Anhydrase (cluster #2 Of 3), Fungal |
Fungi |
110 |
0.44 |
Binding ≤ 10μM
|
|
Q3I4V7-4-F |
Carbonic Anhydrase 2 (cluster #4 Of 4), Fungal |
Fungi |
370 |
0.41 |
Binding ≤ 10μM
|
|
Q5AJ71-1-F |
Carbonic Anhydrase (cluster #1 Of 4), Fungal |
Fungi |
1110 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.16 |
-1.52 |
-44.79 |
1 |
9 |
-1 |
113 |
338.358 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.16 |
-2 |
-12.26 |
2 |
9 |
0 |
116 |
339.366 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
1 |
1.05 |
Binding ≤ 10μM
|
|
CAH9-6-E |
Carbonic Anhydrase IX (cluster #6 Of 11), Eukaryotic |
Eukaryotes |
18 |
0.90 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
0.39 |
-10.57 |
2 |
4 |
0 |
69 |
207.638 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
480 |
0.68 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
149 |
0.74 |
Binding ≤ 10μM
|
|
CAH9-6-E |
Carbonic Anhydrase IX (cluster #6 Of 11), Eukaryotic |
Eukaryotes |
41 |
0.80 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.51 |
0.29 |
-13.39 |
2 |
5 |
0 |
93 |
198.203 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
2 |
1.01 |
Binding ≤ 10μM
|
|
CAH9-6-E |
Carbonic Anhydrase IX (cluster #6 Of 11), Eukaryotic |
Eukaryotes |
59 |
0.84 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
0.56 |
-11.18 |
2 |
4 |
0 |
69 |
187.22 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
2 |
1.11 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
36 |
0.95 |
Binding ≤ 10μM
|
|
CAH9-6-E |
Carbonic Anhydrase IX (cluster #6 Of 11), Eukaryotic |
Eukaryotes |
63 |
0.92 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
-0.12 |
-11.55 |
2 |
4 |
0 |
69 |
173.193 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
400 |
0.34 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
13 |
0.42 |
Binding ≤ 10μM
|
|
CAH9-6-E |
Carbonic Anhydrase IX (cluster #6 Of 11), Eukaryotic |
Eukaryotes |
65 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.71 |
2.14 |
-15.46 |
3 |
6 |
0 |
107 |
383.51 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
710 |
0.86 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
58 |
1.01 |
Binding ≤ 10μM
|
|
CAH9-6-E |
Carbonic Anhydrase IX (cluster #6 Of 11), Eukaryotic |
Eukaryotes |
126 |
0.97 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.00 |
-0.49 |
-10.04 |
2 |
4 |
0 |
69 |
167.23 |
5 |
↓
|
|