|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
-2.16 |
-52.34 |
3 |
7 |
1 |
89 |
412.558 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
-3.23 |
-54.88 |
3 |
8 |
1 |
102 |
467.619 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATC-2-E |
Dipeptidyl Peptidase I (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Binding ≤ 10μM |
|
CATK-1-E |
Cathepsin K (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM |
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
501 |
0.44 |
Binding ≤ 10μM |
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.42 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
5.93 |
-11.49 |
1 |
5 |
0 |
73 |
423.13 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.80 |
6.13 |
-33.48 |
1 |
5 |
0 |
76 |
423.13 |
3 |
↓
|
|
|
|
Analogs
-
4475165
-
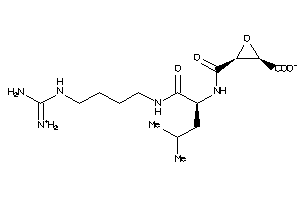
-
13493525
-
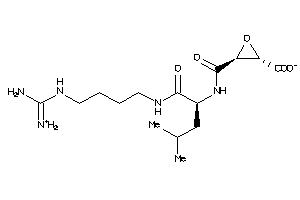
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
16 |
0.44 |
Binding ≤ 10μM
|
|
CATC-1-E |
Dipeptidyl Peptidase I (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
770 |
0.34 |
Binding ≤ 10μM
|
|
CATH-1-E |
Cathepsin H (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
PAPA1-2-E |
Papain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.41 |
Binding ≤ 10μM
|
|
Q9NBA7-1-E |
Cysteine Protease Falcipain-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
75 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
9300 |
0.28 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
2500 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.37 |
1.58 |
-72.44 |
7 |
10 |
0 |
174 |
357.411 |
12 |
↓
|
|
|
|
Analogs
-
13493525
-

-
4095645
-
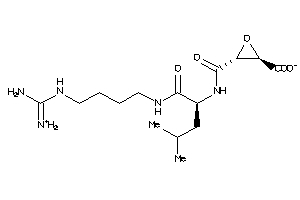
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
16 |
0.44 |
Binding ≤ 10μM
|
|
CATC-1-E |
Dipeptidyl Peptidase I (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
770 |
0.34 |
Binding ≤ 10μM
|
|
CATH-1-E |
Cathepsin H (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
PAPA1-2-E |
Papain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.41 |
Binding ≤ 10μM
|
|
Q9NBA7-1-E |
Cysteine Protease Falcipain-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
75 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
9300 |
0.28 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
2500 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.37 |
1.94 |
-73.1 |
7 |
10 |
0 |
174 |
357.411 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
5.18 |
-13.56 |
2 |
6 |
0 |
91 |
303.362 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.80 |
2.8 |
-43.27 |
1 |
6 |
-1 |
98 |
302.354 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.61 |
4.36 |
-10.14 |
2 |
6 |
0 |
91 |
289.335 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6200 |
0.20 |
Binding ≤ 10μM
|
|
CATF-1-E |
Cathepsin F (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.23 |
Binding ≤ 10μM
|
|
CATK-1-E |
Cathepsin K (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.28 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
760 |
0.23 |
Binding ≤ 10μM
|
|
CATL2-1-E |
Cathepsin L2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
75 |
0.27 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.26 |
Binding ≤ 10μM
|
|
CMA1-1-E |
Mast Cell Protease 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.28 |
Binding ≤ 10μM
|
|
Z50643-1-O |
Hepatitis C Virus (cluster #1 Of 2), Other |
Other |
14 |
0.30 |
Binding ≤ 10μM
|
|
Z50643-5-O |
Hepatitis C Virus (cluster #5 Of 5), Other |
Other |
200 |
0.25 |
Functional ≤ 10μM |
|
A3EZI9-2-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #2 Of 3), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
|
A3EZI9-2-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #2 Of 3), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
|
D2K2A8-1-V |
Hepatitis C Virus NS4A Protein (cluster #1 Of 1), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
6.12 |
-12.14 |
5 |
10 |
0 |
151 |
519.687 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6200 |
0.20 |
Binding ≤ 10μM
|
|
CATF-1-E |
Cathepsin F (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.23 |
Binding ≤ 10μM
|
|
CATK-1-E |
Cathepsin K (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.28 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
760 |
0.23 |
Binding ≤ 10μM
|
|
CATL2-1-E |
Cathepsin L2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
75 |
0.27 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.26 |
Binding ≤ 10μM
|
|
CMA1-1-E |
Mast Cell Protease 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.28 |
Binding ≤ 10μM
|
|
Z50643-1-O |
Hepatitis C Virus (cluster #1 Of 2), Other |
Other |
14 |
0.30 |
Binding ≤ 10μM
|
|
Z50643-5-O |
Hepatitis C Virus (cluster #5 Of 5), Other |
Other |
200 |
0.25 |
Functional ≤ 10μM |
|
A3EZI9-2-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #2 Of 3), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
6.18 |
-13.07 |
5 |
10 |
0 |
151 |
519.687 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4400 |
0.15 |
Binding ≤ 10μM
|
|
CATK-1-E |
Cathepsin K (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
630 |
0.18 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3500 |
0.16 |
Binding ≤ 10μM
|
|
CATL2-1-E |
Cathepsin L2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1350 |
0.17 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.19 |
Binding ≤ 10μM
|
|
CELA1-1-E |
Elastase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.21 |
Binding ≤ 10μM
|
|
CMA1-1-E |
Mast Cell Protease 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.22 |
Binding ≤ 10μM
|
|
ELNE-2-E |
Neutrophil Elastase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8000 |
0.15 |
Binding ≤ 10μM
|
|
Z50643-1-O |
Hepatitis C Virus (cluster #1 Of 2), Other |
Other |
44 |
0.21 |
Binding ≤ 10μM
|
|
Z50643-5-O |
Hepatitis C Virus (cluster #5 Of 5), Other |
Other |
9600 |
0.14 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
8.42 |
-16.9 |
4 |
13 |
0 |
180 |
679.863 |
14 |
↓
|
|
|
|
Analogs
-
2369677
-
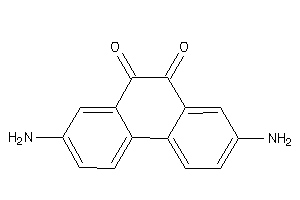
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
5.46 |
-11.78 |
2 |
3 |
0 |
60 |
223.231 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
8.66 |
-25.86 |
2 |
6 |
0 |
99 |
525.568 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.64 |
8.51 |
-21.23 |
1 |
4 |
0 |
63 |
307.349 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
7.43 |
-16.98 |
2 |
7 |
0 |
100 |
401.898 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.29 |
5.07 |
-42.14 |
1 |
7 |
-1 |
106 |
400.89 |
7 |
↓
|
|