|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.28 |
Binding ≤ 10μM
|
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4250 |
0.18 |
Binding ≤ 10μM |
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.29 |
Binding ≤ 10μM |
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.28 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.18 |
-9.49 |
-86.16 |
9 |
12 |
0 |
213 |
596.71 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.16 |
Binding ≤ 10μM |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.16 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.25 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.22 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.26 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
2600 |
0.16 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.5 |
-66.52 |
7 |
14 |
-1 |
225 |
695.819 |
19 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.56 |
5.52 |
-29.09 |
8 |
14 |
0 |
222 |
696.827 |
19 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
146 |
0.25 |
Binding ≤ 10μM |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
146 |
0.25 |
Binding ≤ 10μM |
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM |
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.16 |
12.94 |
-21.59 |
2 |
7 |
0 |
91 |
516.601 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.23 |
Binding ≤ 10μM
|
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
315 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.75 |
18.94 |
-29.57 |
1 |
8 |
0 |
87 |
594.719 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
130 |
0.32 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
48 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
8.34 |
-17.73 |
2 |
6 |
0 |
74 |
463.335 |
3 |
↓
|
|
|
|
Analogs
-
13732965
-
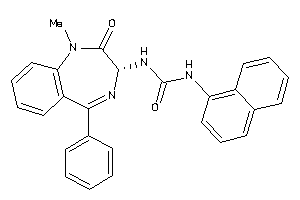
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
68 |
0.30 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
79 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
10.15 |
-19.08 |
2 |
6 |
0 |
74 |
434.499 |
3 |
↓
|
|
|
|
Analogs
-
13732963
-
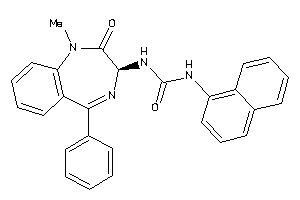
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
68 |
0.30 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
79 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
10.07 |
-15.33 |
2 |
6 |
0 |
74 |
434.499 |
3 |
↓
|
|
|
|
Analogs
-
13732795
-
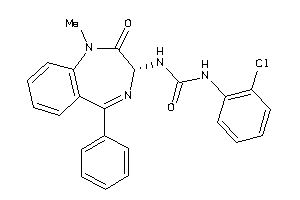
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
410 |
0.30 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
61 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.82 |
8.27 |
-13.68 |
2 |
6 |
0 |
74 |
418.884 |
3 |
↓
|
|
|
|
Analogs
-
13732792
-
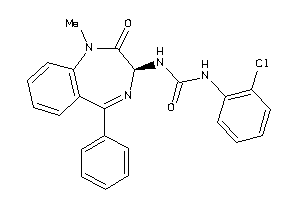
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
410 |
0.30 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
61 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.82 |
8.23 |
-13.58 |
2 |
6 |
0 |
74 |
418.884 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.27 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
7.54 |
-18.31 |
2 |
6 |
0 |
74 |
390.487 |
3 |
↓
|
|
|
|
Analogs
-
13732808
-
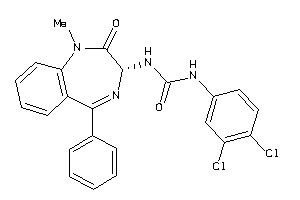
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.34 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
67 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
8.7 |
-19.66 |
2 |
6 |
0 |
74 |
453.329 |
3 |
↓
|
|
|
|
Analogs
-
13732805
-
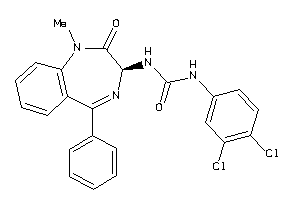
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.34 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
67 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
8.58 |
-12.44 |
2 |
6 |
0 |
74 |
453.329 |
3 |
↓
|
|
|
|
Analogs
-
13732756
-
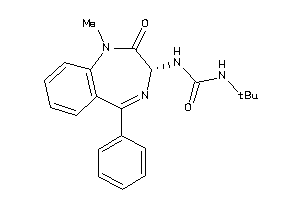
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6800 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
6.05 |
-17.56 |
2 |
6 |
0 |
74 |
364.449 |
3 |
↓
|
|
|
|
Analogs
-
13732753
-
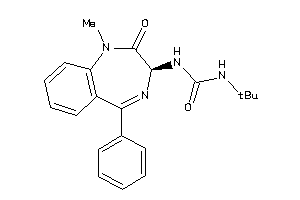
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6800 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
5.97 |
-13.74 |
2 |
6 |
0 |
74 |
364.449 |
3 |
↓
|
|
|
|
Analogs
-
27427028
-
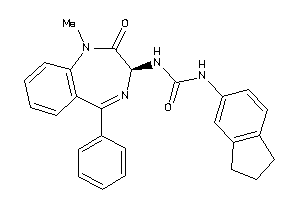
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
900 |
0.26 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
9.54 |
-15.04 |
2 |
6 |
0 |
74 |
424.504 |
3 |
↓
|
|
|
|
Analogs
-
13732897
-
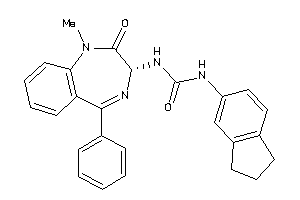
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
900 |
0.26 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
9.5 |
-17.24 |
2 |
6 |
0 |
74 |
424.504 |
3 |
↓
|
|
|
|
Analogs
-
27430688
-
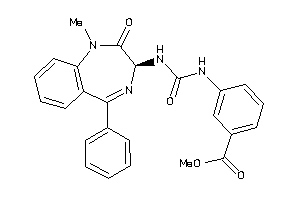
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.25 |
Binding ≤ 10μM
|
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.25 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
8.28 |
-21.9 |
2 |
8 |
0 |
100 |
442.475 |
5 |
↓
|
|
|
|
Analogs
-
13732955
-
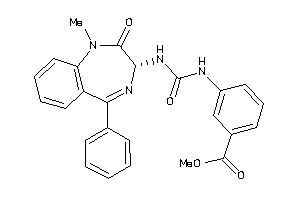
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.25 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
8.33 |
-30.86 |
2 |
8 |
0 |
100 |
442.475 |
5 |
↓
|
|
|
|
Analogs
-
13732790
-
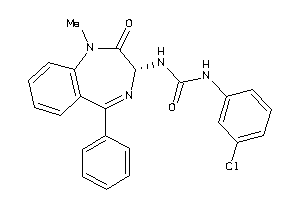
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.38 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
8.16 |
-15.71 |
2 |
6 |
0 |
74 |
418.884 |
3 |
↓
|
|
|
|
Analogs
-
13732787
-
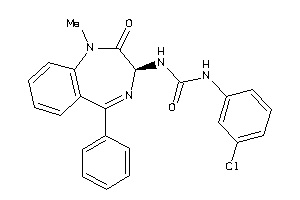
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.38 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
8.17 |
-15.59 |
2 |
6 |
0 |
74 |
418.884 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
490 |
0.29 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
7.74 |
-14.22 |
2 |
6 |
0 |
74 |
398.466 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
210 |
0.29 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
46 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
8.39 |
-20.76 |
2 |
9 |
0 |
120 |
429.436 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.25 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
8.04 |
-27.28 |
2 |
7 |
0 |
98 |
409.449 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
9.99 |
-24.3 |
1 |
5 |
1 |
56 |
414.46 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.08 |
9.39 |
-16.77 |
0 |
5 |
0 |
55 |
413.452 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
9.98 |
-24.27 |
1 |
5 |
1 |
56 |
414.46 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.08 |
9.38 |
-16.81 |
0 |
5 |
0 |
55 |
413.452 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.16 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.97 |
-61.61 |
7 |
14 |
-1 |
225 |
695.819 |
19 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.56 |
5.99 |
-25.52 |
8 |
14 |
0 |
222 |
696.827 |
19 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.16 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.48 |
-63.4 |
7 |
14 |
-1 |
225 |
695.819 |
19 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.56 |
5.5 |
-28.69 |
8 |
14 |
0 |
222 |
696.827 |
19 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.16 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.94 |
-62.59 |
7 |
14 |
-1 |
225 |
695.819 |
19 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.56 |
5.97 |
-26.9 |
8 |
14 |
0 |
222 |
696.827 |
19 |
↓
|
|
|
|
Analogs
-
13732781
-
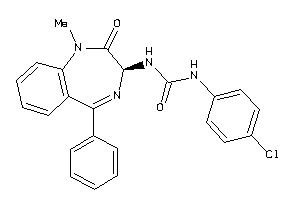
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
51 |
0.34 |
Binding ≤ 10μM
|
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.38 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
81 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.87 |
8.2 |
-18.07 |
2 |
6 |
0 |
74 |
418.884 |
3 |
↓
|
|
|
|
Analogs
-
1532369
-
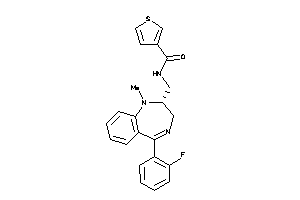
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
47 |
0.37 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
153 |
0.34 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
-3.67 |
-14.18 |
1 |
4 |
0 |
44 |
393.487 |
4 |
↓
|
|
|
|
Analogs
-
967961
-
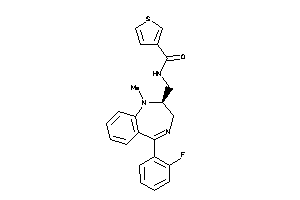
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
47 |
0.37 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
153 |
0.34 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
-3.23 |
-16.69 |
1 |
4 |
0 |
44 |
393.487 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
197 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
-1.48 |
-60.03 |
3 |
7 |
-1 |
114 |
426.452 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
-0.51 |
-15.82 |
1 |
4 |
0 |
48 |
397.453 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
10.25 |
-18.65 |
1 |
4 |
0 |
48 |
397.453 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.14 |
10.77 |
-28.23 |
2 |
4 |
1 |
50 |
398.461 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
66 |
0.34 |
Binding ≤ 10μM
|
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.27 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.26 |
Binding ≤ 10μM |
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
180 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.43 |
12.58 |
-52.21 |
1 |
6 |
-1 |
90 |
458.406 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Binding ≤ 10μM
|
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.25 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.43 |
12.64 |
-50.35 |
1 |
6 |
-1 |
90 |
458.406 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.17 |
Binding ≤ 10μM |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.17 |
Binding ≤ 10μM |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.17 |
Binding ≤ 10μM |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.17 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.27 |
Binding ≤ 10μM |
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.27 |
Binding ≤ 10μM |
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
63 |
0.22 |
Binding ≤ 10μM |
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.17 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.69 |
-4.42 |
-53.62 |
4 |
10 |
-1 |
152 |
613.735 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
502 |
0.24 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
-4.12 |
-20.59 |
3 |
9 |
0 |
115 |
498.587 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
34 |
0.33 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
8.39 |
-17.77 |
2 |
9 |
0 |
120 |
429.436 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.37 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
54 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
7.71 |
-16.71 |
2 |
6 |
0 |
74 |
402.429 |
3 |
↓
|
|
|
|
Analogs
-
13732802
-
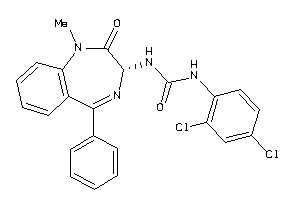
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.38 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
8.86 |
-14.02 |
2 |
6 |
0 |
74 |
453.329 |
3 |
↓
|
|
|
|
Analogs
-
13732799
-
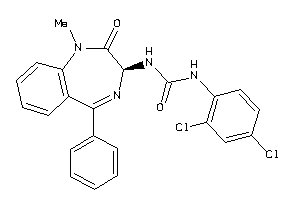
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.38 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
8.74 |
-11.5 |
2 |
6 |
0 |
74 |
453.329 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.26 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
8.05 |
-32.56 |
2 |
7 |
0 |
91 |
426.476 |
4 |
↓
|
|
|
|
Analogs
-
16740073
-
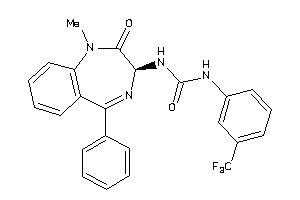
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.26 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
8.65 |
-13.18 |
2 |
6 |
0 |
74 |
452.436 |
4 |
↓
|
|
|
|
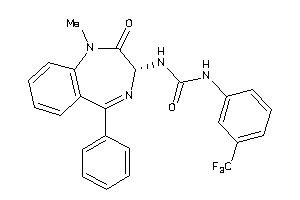
Analogs
-
13732894
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.26 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
8.73 |
-16.54 |
2 |
6 |
0 |
74 |
452.436 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
146 |
0.25 |
Binding ≤ 10μM |
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.16 |
13.01 |
-24.49 |
2 |
7 |
0 |
91 |
516.601 |
6 |
↓
|
|
|
|
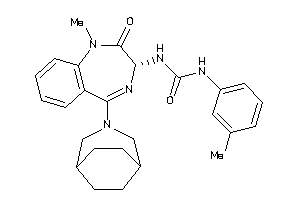
Analogs
-
49625618
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.35 |
Binding ≤ 10μM
|
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.33 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
9.7 |
-15.49 |
2 |
7 |
0 |
77 |
445.567 |
3 |
↓
|
|