|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNH-1-E |
Cyclin H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.33 |
Binding ≤ 10μM |
|
CCNT1-1-E |
Cyclin T1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
90 |
0.35 |
Binding ≤ 10μM |
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.38 |
Binding ≤ 10μM |
|
CDK7-1-E |
Cyclin-dependent Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.33 |
Binding ≤ 10μM |
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.35 |
Binding ≤ 10μM |
|
DYR1A-3-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2100 |
0.28 |
Binding ≤ 10μM |
|
INSRR-1-E |
Insulin Receptor-related Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.29 |
Binding ≤ 10μM |
|
KC1A-1-E |
Casein Kinase I Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1040 |
0.30 |
Binding ≤ 10μM |
|
KKCC2-1-E |
CaM-kinase Kinase Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2450 |
0.28 |
Binding ≤ 10μM |
|
MAT1-1-E |
CDK-activating Kinase Assembly Factor MAT1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.33 |
Binding ≤ 10μM |
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3110 |
0.28 |
Binding ≤ 10μM |
|
MK03-1-E |
MAP Kinase ERK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3730 |
0.27 |
Binding ≤ 10μM |
|
MK15-1-E |
Mitogen-activated Protein Kinase 15 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
330 |
0.32 |
Binding ≤ 10μM |
|
Z104296-1-O |
Cyclin-dependent Kinase 1/cyclin B1 (cluster #1 Of 2), Other |
Other |
33 |
0.37 |
Binding ≤ 10μM |
|
Z104299-1-O |
Cyclin-dependent Kinase 2/cyclin E1 (cluster #1 Of 1), Other |
Other |
3 |
0.43 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
1.79 |
-12.19 |
5 |
8 |
0 |
115 |
385.468 |
9 |
↓
|
|
|
|
Analogs
-
602192
-
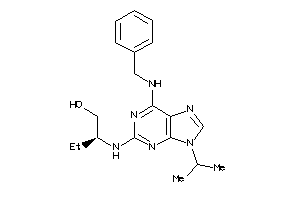
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.30 |
Binding ≤ 10μM
|
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNH-1-E |
Cyclin H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
540 |
0.34 |
Binding ≤ 10μM |
|
CCNT1-1-E |
Cyclin T1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
950 |
0.32 |
Binding ≤ 10μM |
|
CCNT2-1-E |
Cyclin-T2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
810 |
0.33 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CDK16-1-E |
Serine/threonine-protein Kinase PCTAIRE-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
990 |
0.32 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CDK4-2-E |
Cyclin-dependent Kinase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.38 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.37 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.35 |
Binding ≤ 10μM
|
|
CDK7-1-E |
Cyclin-dependent Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.33 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
950 |
0.32 |
Binding ≤ 10μM |
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.31 |
Binding ≤ 10μM
|
|
CLK2-1-E |
Dual Specificity Protein Kinase CLK2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CLK4-1-E |
Dual Specificity Protein Kinase CLK4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.29 |
Binding ≤ 10μM
|
|
DYR1A-3-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.29 |
Binding ≤ 10μM
|
|
DYR1B-1-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.32 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
260 |
0.35 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.35 |
Binding ≤ 10μM
|
|
KC1G1-1-E |
Casein Kinase I Gamma 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.30 |
Binding ≤ 10μM
|
|
KC1G2-1-E |
Casein Kinase I Gamma 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.32 |
Binding ≤ 10μM
|
|
KC1G3-1-E |
Casein Kinase I Isoform Gamma-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.30 |
Binding ≤ 10μM
|
|
KS6A2-1-E |
Ribosomal Protein S6 Kinase Alpha 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.30 |
Binding ≤ 10μM
|
|
MAT1-1-E |
CDK-activating Kinase Assembly Factor MAT1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
540 |
0.34 |
Binding ≤ 10μM |
|
PAK4-1-E |
Serine/threonine-protein Kinase PAK 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.28 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.31 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
280 |
0.35 |
Binding ≤ 10μM |
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
940 |
0.32 |
Binding ≤ 10μM
|
|
Z104296-2-O |
Cyclin-dependent Kinase 1/cyclin B1 (cluster #2 Of 2), Other |
Other |
450 |
0.34 |
Binding ≤ 10μM
|
|
Z104299-1-O |
Cyclin-dependent Kinase 2/cyclin E1 (cluster #1 Of 1), Other |
Other |
220 |
0.36 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
6310 |
0.28 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
6000 |
0.28 |
Functional ≤ 10μM
|
|
Z80390-10-O |
PC-3 (Prostate Carcinoma Cells) (cluster #10 Of 10), Other |
Other |
8000 |
0.27 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
5000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
9.65 |
-10.26 |
3 |
7 |
0 |
88 |
354.458 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.65 |
9.9 |
-28.83 |
4 |
7 |
1 |
89 |
355.466 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.65 |
9.47 |
-25.05 |
4 |
7 |
1 |
89 |
355.466 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.17 |
-2.68 |
-9.08 |
6 |
7 |
0 |
126 |
218.22 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.17 |
-1.43 |
-10.95 |
6 |
7 |
0 |
126 |
218.22 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.17 |
-1.91 |
-46.56 |
5 |
7 |
-1 |
129 |
217.212 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.62 |
Binding ≤ 10μM
|
|
CCNT1-1-E |
Cyclin T1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.64 |
Binding ≤ 10μM
|
|
CDK2-5-E |
Cyclin-dependent Kinase 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
11 |
0.62 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.64 |
Binding ≤ 10μM
|
|
DYR1A-1-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
29 |
0.59 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
170 |
0.53 |
Binding ≤ 10μM
|
|
Z80140-1-O |
GBM (cluster #1 Of 1), Other |
Other |
140 |
0.53 |
Functional ≤ 10μM
|
|
Z80928-2-O |
HCT-116 (Colon Carcinoma Cells) (cluster #2 Of 9), Other |
Other |
63 |
0.56 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
240 |
0.51 |
Functional ≤ 10μM
|
|
Z81186-2-O |
LS174T (Colon Adencocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
130 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.72 |
4.85 |
-14.34 |
3 |
6 |
0 |
90 |
241.254 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.72 |
5.12 |
-39.43 |
4 |
6 |
1 |
91 |
242.262 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
6.73 |
-13.97 |
3 |
6 |
0 |
90 |
269.308 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.60 |
7 |
-39.58 |
4 |
6 |
1 |
91 |
270.316 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1300 |
0.48 |
Binding ≤ 10μM
|
|
CCNT1-1-E |
Cyclin T1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Binding ≤ 10μM
|
|
CDK2-5-E |
Cyclin-dependent Kinase 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
1300 |
0.48 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Binding ≤ 10μM
|
|
DYR1A-1-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.54 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
680 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
5.35 |
-13.79 |
3 |
5 |
0 |
80 |
290.124 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
6 |
-14.2 |
3 |
5 |
0 |
80 |
245.673 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
780 |
0.43 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
CCNT1-1-E |
Cyclin T1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
64 |
0.50 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
780 |
0.43 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
64 |
0.50 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.53 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
230 |
0.46 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
470 |
0.44 |
Binding ≤ 10μM
|
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.35 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
850 |
0.42 |
Binding ≤ 10μM
|
|
Z104296-1-O |
Cyclin-dependent Kinase 1/cyclin B1 (cluster #1 Of 2), Other |
Other |
400 |
0.45 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
4000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.72 |
7.21 |
-9.5 |
2 |
3 |
0 |
45 |
327.181 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.41 |
Binding ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.41 |
Binding ≤ 10μM
|
|
AURKA-3-E |
Serine/threonine-protein Kinase Aurora-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.43 |
Binding ≤ 10μM
|
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
57 |
0.44 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.43 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.43 |
Binding ≤ 10μM
|
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
CCNH-1-E |
Cyclin H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.44 |
Binding ≤ 10μM
|
|
CCNT1-1-E |
Cyclin T1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.43 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
CDK4-1-E |
Cyclin-dependent Kinase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
53 |
0.44 |
Binding ≤ 10μM
|
|
CDK7-1-E |
Cyclin-dependent Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.44 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
20 |
0.47 |
Binding ≤ 10μM
|
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
53 |
0.44 |
Binding ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
300 |
0.40 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
48 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
-2.06 |
-14.33 |
3 |
8 |
0 |
122 |
328.357 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.41 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Binding ≤ 10μM
|
|
CCNT1-1-E |
Cyclin T1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
810 |
0.45 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2700 |
0.41 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
810 |
0.45 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
620 |
0.46 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
4200 |
0.40 |
Binding ≤ 10μM
|
|
Z104296-1-O |
Cyclin-dependent Kinase 1/cyclin B1 (cluster #1 Of 2), Other |
Other |
7000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
-1.4 |
-9.56 |
2 |
3 |
0 |
44 |
248.285 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-3-E |
Serine/threonine-protein Kinase Aurora-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.44 |
Binding ≤ 10μM
|
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.43 |
Binding ≤ 10μM
|
|
AURKC-1-E |
Serine/threonine-protein Kinase Aurora-C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
65 |
0.39 |
Binding ≤ 10μM
|
|
CCNT1-1-E |
Cyclin T1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
480 |
0.34 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
390 |
0.35 |
Binding ≤ 10μM
|
|
CDK4-1-E |
Cyclin-dependent Kinase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1090 |
0.32 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
480 |
0.34 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
88 |
0.38 |
Binding ≤ 10μM
|
|
KS6B1-1-E |
Ribosomal Protein S6 Kinase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
540 |
0.34 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2800 |
0.30 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
820 |
0.33 |
Binding ≤ 10μM
|
|
VGFR2-2-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
69 |
0.39 |
Binding ≤ 10μM
|
|
Z104299-1-O |
Cyclin-dependent Kinase 2/cyclin E1 (cluster #1 Of 1), Other |
Other |
390 |
0.35 |
Binding ≤ 10μM
|
|
Z100405-1-O |
MV4-11 (Myeloid Leukemia Cells) (cluster #1 Of 1), Other |
Other |
34 |
0.40 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
471 |
0.34 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
372 |
0.35 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
725 |
0.33 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
1375 |
0.32 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
599 |
0.34 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
340 |
0.35 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
681 |
0.33 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
170 |
0.36 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
590 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
-5.83 |
-10.66 |
3 |
7 |
0 |
89 |
368.466 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.40 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
16 |
0.42 |
Binding ≤ 10μM
|
|
CCND3-1-E |
G1/S-specific Cyclin D3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
21 |
0.41 |
Binding ≤ 10μM
|
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
CCNT1-1-E |
Cyclin T1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.41 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
62 |
0.39 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
CDK4-1-E |
Cyclin-dependent Kinase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.34 |
Binding ≤ 10μM
|
|
KCNH2-2-E |
HERG (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
3200 |
0.30 |
Binding ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
800 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.63 |
8.32 |
-41.84 |
2 |
7 |
1 |
91 |
372.474 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.63 |
7.85 |
-19.57 |
1 |
7 |
0 |
90 |
371.466 |
5 |
↓
|
|