|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.61 |
2.85 |
-12.66 |
3 |
3 |
0 |
44 |
264.272 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2500 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.38 |
3.89 |
-15.44 |
2 |
2 |
0 |
32 |
282.217 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2530 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
0.09 |
-9.3 |
3 |
4 |
0 |
70 |
229.663 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3230 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.08 |
-0.52 |
-40.08 |
4 |
5 |
1 |
78 |
297.762 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.08 |
-0.23 |
-42.85 |
2 |
5 |
-1 |
80 |
295.746 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.08 |
0.03 |
-86.69 |
5 |
5 |
2 |
79 |
298.77 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
830 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
-2.86 |
-83.94 |
4 |
4 |
2 |
50 |
309.841 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4900 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.38 |
0.96 |
-15.43 |
2 |
4 |
0 |
74 |
326.392 |
9 |
↓
|
|
|
|
Analogs
-
11616527
-
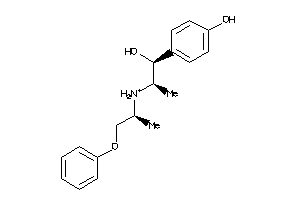
-
3201958
-
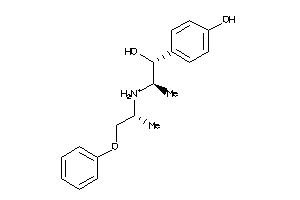
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2900 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
4.31 |
-41.46 |
4 |
4 |
1 |
66 |
302.394 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.77 |
3.61 |
-6.35 |
3 |
4 |
0 |
62 |
301.386 |
7 |
↓
|
|
|
|
Analogs
-
31911781
-
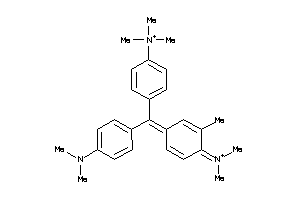
-
3953819
-
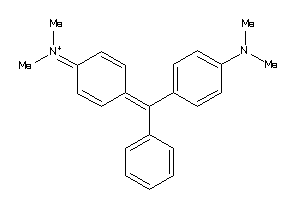
Draw
Identity
99%
90%
80%
70%
Vendors
And 48 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
Z104295-2-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #2 Of 2), Other |
Other |
590 |
0.31 |
Binding ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
2100 |
0.28 |
Functional ≤ 10μM
|
|
Z80021-4-O |
A498 (Renal Carcinoma Cells) (cluster #4 Of 5), Other |
Other |
57 |
0.36 |
Functional ≤ 10μM
|
|
Z80099-3-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
13 |
0.39 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
15 |
0.39 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
14 |
0.39 |
Functional ≤ 10μM
|
|
Z80712-4-O |
T47D (Breast Carcinoma Cells) (cluster #4 Of 7), Other |
Other |
16 |
0.39 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
34 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
13.34 |
-19.69 |
0 |
3 |
1 |
9 |
372.536 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.42 |
Binding ≤ 10μM
|
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
169 |
0.38 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.42 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
169 |
0.38 |
Binding ≤ 10μM
|
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
208 |
0.37 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
208 |
0.37 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
208 |
0.37 |
Binding ≤ 10μM
|
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
55 |
0.41 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
55 |
0.41 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
208 |
0.37 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
55 |
0.41 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
169 |
0.38 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
732 |
0.34 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
65 |
0.40 |
Binding ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
639 |
0.35 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
1354 |
0.33 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
630 |
0.35 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
800 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
8.37 |
-18.97 |
2 |
6 |
0 |
78 |
338.411 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
2.17 |
8.31 |
-20.43 |
2 |
6 |
0 |
78 |
338.411 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB-1-E |
G2/mitotic-specific Cyclin B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.23 |
Binding ≤ 10μM |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.23 |
Binding ≤ 10μM |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
2930 |
0.25 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
10.79 |
-15.5 |
3 |
7 |
0 |
96 |
406.449 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.76 |
8.46 |
-8.6 |
3 |
7 |
0 |
99 |
406.449 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.76 |
9.65 |
-34.29 |
4 |
7 |
1 |
101 |
407.457 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.29 |
Binding ≤ 10μM
|
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
|
AURKA-2-E |
Serine/threonine-protein Kinase Aurora-A (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
760 |
0.31 |
Binding ≤ 10μM |
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
390 |
0.32 |
Binding ≤ 10μM
|
|
AURKC-2-E |
Serine/threonine-protein Kinase Aurora-C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
71 |
0.36 |
Binding ≤ 10μM
|
|
BMPR2-1-E |
Bone Morphogenetic Protein Receptor Type-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
CD11A-1-E |
PITSLRE Serine/threonine-protein Kinase CDC2L2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.30 |
Binding ≤ 10μM
|
|
CD11B-1-E |
PITSLRE Serine/threonine-protein Kinase CDC2L1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.29 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
9800 |
0.25 |
Binding ≤ 10μM
|
|
CDK19-1-E |
Cell Division Cycle 2-like Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
74 |
0.36 |
Binding ≤ 10μM
|
|
CDK7-1-E |
Cyclin-dependent Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.28 |
Binding ≤ 10μM
|
|
CDK8-1-E |
Cell Division Protein Kinase 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
95 |
0.35 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.28 |
Binding ≤ 10μM
|
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8900 |
0.25 |
Binding ≤ 10μM
|
|
CLK4-2-E |
Dual Specificity Protein Kinase CLK4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
58 |
0.36 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3800 |
0.27 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6500 |
0.26 |
Binding ≤ 10μM
|
|
EPHA3-1-E |
Ephrin Type-A Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.27 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
330 |
0.32 |
Binding ≤ 10μM
|
|
EPHB3-1-E |
Ephrin Type-B Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.29 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.29 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.32 |
Binding ≤ 10μM
|
|
INSRR-1-E |
Insulin Receptor-related Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3900 |
0.27 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6900 |
0.26 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
81 |
0.35 |
Binding ≤ 10μM
|
|
KS6A4-1-E |
Ribosomal Protein S6 Kinase Alpha 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.26 |
Binding ≤ 10μM
|
|
KS6A5-1-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.30 |
Binding ≤ 10μM
|
|
LATS2-1-E |
Serine/threonine-protein Kinase LATS2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.29 |
Binding ≤ 10μM
|
|
LIMK2-1-E |
LIM Domain Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.27 |
Binding ≤ 10μM
|
|
LTK-1-E |
Leukocyte Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
550 |
0.31 |
Binding ≤ 10μM
|
|
M4K1-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
830 |
0.30 |
Binding ≤ 10μM
|
|
M4K3-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.27 |
Binding ≤ 10μM
|
|
M4K5-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
910 |
0.30 |
Binding ≤ 10μM
|
|
MERTK-1-E |
Proto-oncogene Tyrosine-protein Kinase MER (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
870 |
0.30 |
Binding ≤ 10μM
|
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.29 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.33 |
Binding ≤ 10μM
|
|
MK15-1-E |
Mitogen-activated Protein Kinase 15 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.27 |
Binding ≤ 10μM
|
|
MKNK2-1-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
270 |
0.33 |
Binding ≤ 10μM
|
|
MUSK-1-E |
Muscle, Skeletal Receptor Tyrosine Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
NTRK1-1-E |
Nerve Growth Factor Receptor Trk-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.28 |
Binding ≤ 10μM
|
|
NTRK2-1-E |
Neurotrophic Tyrosine Kinase Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.30 |
Binding ≤ 10μM
|
|
NTRK3-1-E |
NT-3 Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
210 |
0.33 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
66 |
0.36 |
Binding ≤ 10μM
|
|
PLK3-1-E |
Serine/threonine-protein Kinase PLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8500 |
0.25 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.29 |
Binding ≤ 10μM
|
|
RIPK1-1-E |
Receptor-interacting Serine/threonine-protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
450 |
0.32 |
Binding ≤ 10μM
|
|
ST32B-1-E |
Serine/threonine-protein Kinase 32B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.33 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.32 |
Binding ≤ 10μM
|
|
STK3-1-E |
Serine/threonine-protein Kinase MST2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.29 |
Binding ≤ 10μM
|
|
STK33-1-E |
Serine/threonine-protein Kinase 33 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3800 |
0.27 |
Binding ≤ 10μM
|
|
STK4-2-E |
Serine/threonine-protein Kinase MST1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.29 |
Binding ≤ 10μM
|
|
TIE1-1-E |
Tyrosine-protein Kinase Receptor Tie-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.35 |
Binding ≤ 10μM
|
|
TIE2-1-E |
Tyrosine-protein Kinase TIE-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
450 |
0.32 |
Binding ≤ 10μM
|
|
UFO-1-E |
Tyrosine-protein Kinase Receptor UFO (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.32 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.34 |
Binding ≤ 10μM
|
|
VGFR2-2-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.73 |
7.88 |
-12.09 |
5 |
6 |
0 |
96 |
375.407 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2,3-diphenyl-4,5,7,8-tetrazabicyclo[4.3.0]nona-2,4,8,10-tetraen-9-amine
2,3-diphenyl-4,5,7,8-tetrazabicy…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB-1-E |
G2/mitotic-specific Cyclin B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6100 |
0.33 |
Binding ≤ 10μM |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6100 |
0.33 |
Binding ≤ 10μM |
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.42 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
250 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
7.27 |
-8.85 |
3 |
5 |
0 |
80 |
287.326 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM |
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
140 |
0.34 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
140 |
0.34 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5200 |
0.26 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5200 |
0.26 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5200 |
0.26 |
Binding ≤ 10μM
|
|
KC1A-1-E |
Casein Kinase I Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
KC1G1-1-E |
Casein Kinase I Gamma 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
KC1G2-1-E |
Casein Kinase I Gamma 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
KCC2A-1-E |
CaM Kinase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8900 |
0.25 |
Binding ≤ 10μM
|
|
KCC2B-1-E |
CaM Kinase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8900 |
0.25 |
Binding ≤ 10μM
|
|
KCC2D-1-E |
CaM Kinase II Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8900 |
0.25 |
Binding ≤ 10μM
|
|
KCC2G-1-E |
CaM Kinase II Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8900 |
0.25 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
|
Q95J97-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5200 |
0.26 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
32 |
0.37 |
Binding ≤ 10μM
|
|
Z104300-1-O |
Mitogen-activated Protein Kinase; ERK1/ERK2 (cluster #1 Of 1), Other |
Other |
1000 |
0.30 |
Binding ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
92 |
0.35 |
Functional ≤ 10μM
|
|
Z80018-2-O |
A-375 (Malignant Melanoma Cells) (cluster #2 Of 4), Other |
Other |
92 |
0.35 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
20 |
0.38 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
35 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
7.08 |
-16.53 |
6 |
9 |
0 |
141 |
425.446 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.43 |
7.94 |
-52.61 |
5 |
9 |
-1 |
141 |
424.438 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1130 |
0.33 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1130 |
0.33 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1130 |
0.33 |
Binding ≤ 10μM
|
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1130 |
0.33 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
KCC2A-1-E |
CaM Kinase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.43 |
Binding ≤ 10μM
|
|
KCC2B-1-E |
CaM Kinase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.43 |
Binding ≤ 10μM
|
|
KCC2D-1-E |
CaM Kinase II Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.43 |
Binding ≤ 10μM
|
|
KCC2G-1-E |
CaM Kinase II Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.43 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
163 |
0.38 |
Binding ≤ 10μM
|
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
200 |
0.38 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
4000 |
0.30 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
850 |
0.34 |
Functional ≤ 10μM
|
|
Z81024-8-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #8 Of 8), Other |
Other |
590 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
5.67 |
-11.48 |
3 |
5 |
0 |
82 |
325.327 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.35 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.35 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
690 |
0.39 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
690 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
1.4 |
-16.55 |
4 |
6 |
0 |
105 |
315.354 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.70 |
1.3 |
-16.99 |
4 |
6 |
0 |
105 |
315.354 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.22 |
-0.66 |
-14.57 |
4 |
6 |
0 |
109 |
315.354 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
330 |
0.39 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
330 |
0.39 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
46 |
0.45 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
46 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
1.02 |
-17.5 |
4 |
7 |
0 |
117 |
330.369 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
2.12 |
1.14 |
-13.81 |
4 |
7 |
0 |
117 |
330.369 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.30 |
-0.08 |
-50.3 |
3 |
7 |
-1 |
124 |
329.361 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
2300 |
0.53 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
4000 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
-1.21 |
-8.62 |
1 |
3 |
0 |
41 |
195.225 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Binding ≤ 10μM
|
|
CCND1-1-E |
G1/S-specific Cyclin D1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.37 |
Binding ≤ 10μM
|
|
CCND2-1-E |
G1/S-specific Cyclin D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.37 |
Binding ≤ 10μM
|
|
CCND3-1-E |
G1/S-specific Cyclin D3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.37 |
Binding ≤ 10μM
|
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.45 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.45 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Binding ≤ 10μM
|
|
CDK2-5-E |
Cyclin-dependent Kinase 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
9 |
0.45 |
Binding ≤ 10μM
|
|
CDK4-2-E |
Cyclin-dependent Kinase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
230 |
0.37 |
Binding ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
760 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
9.77 |
-10.35 |
1 |
5 |
0 |
68 |
345.349 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.22 |
10.03 |
-34.41 |
2 |
5 |
1 |
69 |
346.357 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.22 |
10.03 |
-35.9 |
2 |
5 |
1 |
69 |
346.357 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-1-E |
Serine/threonine-protein Kinase Aurora-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.35 |
Binding ≤ 10μM
|
|
AURKB-2-E |
Serine/threonine-protein Kinase Aurora-B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2140 |
0.38 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
214 |
0.44 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
140 |
0.46 |
Binding ≤ 10μM
|
|
CDK4-2-E |
Cyclin-dependent Kinase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
3000 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
0.97 |
-11.97 |
3 |
5 |
0 |
83 |
281.267 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
780 |
0.37 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
780 |
0.37 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
60 |
0.44 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
60 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
-1.86 |
-14.06 |
4 |
7 |
0 |
118 |
395.238 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
2.30 |
0.95 |
-40.52 |
5 |
7 |
1 |
115 |
396.246 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.48 |
1.09 |
-13.5 |
4 |
7 |
0 |
117 |
395.238 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
3.26 |
-49.08 |
5 |
7 |
1 |
103 |
383.259 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.48 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.48 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.48 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.48 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.48 |
Binding ≤ 10μM
|
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
150 |
0.48 |
Binding ≤ 10μM
|
|
CDK2-5-E |
Cyclin-dependent Kinase 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.47 |
Binding ≤ 10μM
|
|
CFTR-1-E |
Cystic Fibrosis Transmembrane Conductance Regulator (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.48 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.44 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
650 |
0.43 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3300 |
0.38 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3300 |
0.38 |
Binding ≤ 10μM
|
|
MK10-2-E |
C-Jun N-terminal Kinase 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.38 |
Binding ≤ 10μM
|
|
CDK1-1-F |
Cyclin-dependent Kinase 1 (cluster #1 Of 1), Fungal |
Fungi |
2500 |
0.39 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
200 |
0.47 |
Binding ≤ 10μM
|
|
Z80344-1-O |
NT2 (Teratocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
7000 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
5.32 |
-9.36 |
2 |
4 |
0 |
62 |
267.332 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.78 |
6.16 |
-73.47 |
1 |
4 |
-1 |
60 |
266.324 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.38 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.38 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.38 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3600 |
0.38 |
Binding ≤ 10μM
|
|
CDK2-5-E |
Cyclin-dependent Kinase 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
4500 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
9.66 |
-9.7 |
1 |
5 |
0 |
56 |
267.336 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.41 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.41 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.41 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1200 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
6.72 |
-6.7 |
2 |
5 |
0 |
66 |
267.336 |
4 |
↓
|
|