|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.37 |
Binding ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.37 |
Binding ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
1500 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
6.04 |
-41.95 |
1 |
7 |
-1 |
99 |
317.369 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.60 |
7.03 |
-95.06 |
0 |
7 |
-2 |
102 |
316.361 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.60 |
6.15 |
-17.48 |
2 |
7 |
0 |
101 |
318.377 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.42 |
5.43 |
-10.63 |
1 |
5 |
0 |
68 |
382.363 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.42 |
5.52 |
-36.42 |
0 |
5 |
-1 |
70 |
381.355 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.42 |
5.71 |
-36.98 |
2 |
5 |
1 |
70 |
383.371 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.32 |
9.18 |
-41.96 |
3 |
9 |
1 |
125 |
482.52 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
5.32 |
9.03 |
-36.32 |
3 |
9 |
1 |
125 |
482.52 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.32 |
9.8 |
-16.61 |
2 |
9 |
0 |
127 |
481.512 |
8 |
↓
|
|
|
|
Analogs
-
13741956
-
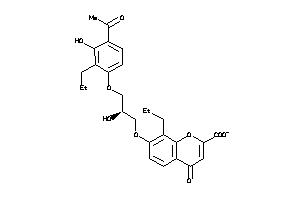
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.23 |
Binding ≤ 10μM
|
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.22 |
Binding ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2007 |
0.22 |
Binding ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.21 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1580 |
0.23 |
Binding ≤ 10μM
|
|
THAS-2-E |
Thromboxane-A Synthase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.19 |
Binding ≤ 10μM
|
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.27 |
Functional ≤ 10μM
|
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.24 |
Functional ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.27 |
Functional ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.24 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
35 |
0.29 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
940 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.77 |
10.26 |
-59.01 |
2 |
9 |
-1 |
146 |
497.52 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.77 |
11.27 |
-106.46 |
1 |
9 |
-2 |
149 |
496.512 |
12 |
↓
|
|
|
|
Analogs
-
13741955
-
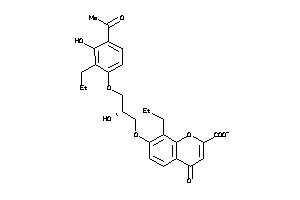
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.23 |
Binding ≤ 10μM
|
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.22 |
Binding ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2007 |
0.22 |
Binding ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.21 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1580 |
0.23 |
Binding ≤ 10μM
|
|
THAS-2-E |
Thromboxane-A Synthase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.19 |
Binding ≤ 10μM
|
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.27 |
Functional ≤ 10μM
|
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.24 |
Functional ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.27 |
Functional ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.24 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
35 |
0.29 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
940 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.77 |
10.26 |
-59.18 |
2 |
9 |
-1 |
146 |
497.52 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.77 |
11.27 |
-106.59 |
1 |
9 |
-2 |
149 |
496.512 |
12 |
↓
|
|
|
|
Analogs
-
34592840
-
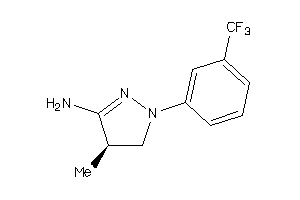
-
34592841
-
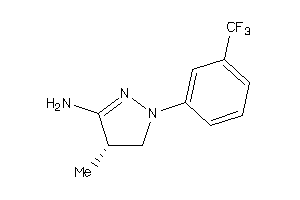
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.54 |
Binding ≤ 10μM
|
|
LOX5-5-E |
Arachidonate 5-lipoxygenase (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
890 |
0.53 |
Binding ≤ 10μM
|
|
LT4R1-1-E |
Leukotriene B4 Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.54 |
Binding ≤ 10μM
|
|
PGH1-3-E |
Cyclooxygenase-1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
800 |
0.53 |
Binding ≤ 10μM |
|
PGH2-3-E |
Cyclooxygenase-2 (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
1300 |
0.52 |
Binding ≤ 10μM |
|
LOX5-3-E |
Arachidonate 5-lipoxygenase (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
750 |
0.54 |
Functional ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.46 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.46 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
8500 |
0.44 |
Functional ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 12), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
1.29 |
-9.17 |
2 |
3 |
0 |
42 |
229.205 |
2 |
↓
|
|
|
|
Analogs
-
4172316
-
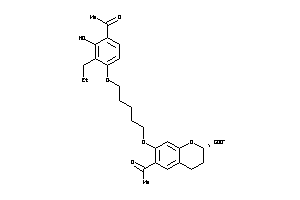
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.21 |
Binding ≤ 10μM
|
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.20 |
Binding ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.20 |
Binding ≤ 10μM
|
|
LT4R1-1-E |
Leukotriene B4 Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.25 |
Binding ≤ 10μM |
|
LT4R2-1-E |
Leukotriene B4 Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.25 |
Binding ≤ 10μM |
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.21 |
Functional ≤ 10μM
|
|
THAS-1-E |
Thromboxane-A Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
60 |
0.28 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
100 |
0.27 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
4000 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.17 |
1.9 |
-59.27 |
1 |
8 |
-1 |
122 |
497.564 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.75 |
9.97 |
-67.37 |
1 |
9 |
-1 |
122 |
574.679 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
5.75 |
9.41 |
-24.1 |
2 |
9 |
0 |
119 |
575.687 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.69 |
12.06 |
-20.43 |
2 |
9 |
0 |
116 |
575.687 |
9 |
↓
|
|
|
|
Analogs
-
11615905
-
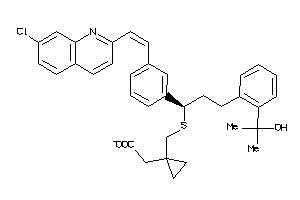
-
11615906
-
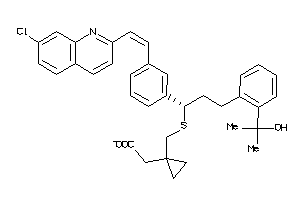
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.30 |
Binding ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.31 |
Binding ≤ 10μM
|
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
72 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.89 |
0.56 |
-56.72 |
1 |
4 |
-1 |
73 |
585.189 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.29 |
Binding ≤ 10μM |
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.34 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
5300 |
0.30 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
5300 |
0.30 |
Binding ≤ 10μM
|
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.35 |
Functional ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.35 |
Functional ≤ 10μM
|
|
LOX5-6-E |
Arachidonate 5-lipoxygenase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
3000 |
0.31 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.29 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
700 |
0.34 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3000 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.28 |
9.63 |
-7.94 |
1 |
3 |
0 |
42 |
335.447 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.29 |
Binding ≤ 10μM |
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.34 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
5300 |
0.30 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
5300 |
0.30 |
Binding ≤ 10μM
|
|
CLTR1-1-E |
Cysteinyl Leukotriene Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.35 |
Functional ≤ 10μM
|
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.35 |
Functional ≤ 10μM
|
|
LOX5-6-E |
Arachidonate 5-lipoxygenase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
3000 |
0.31 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.29 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
700 |
0.34 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3000 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.28 |
9.67 |
-7.89 |
1 |
3 |
0 |
42 |
335.447 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
896731
-
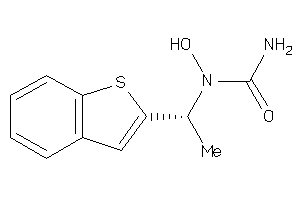
Draw
Identity
99%
90%
80%
70%
Vendors
And 32 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.51 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
830 |
0.53 |
Binding ≤ 10μM
|
|
LT4R1-1-E |
Leukotriene B4 Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
420 |
0.56 |
Binding ≤ 10μM
|
|
LKHA4-1-E |
Leukotriene A4 Hydrolase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
850 |
0.53 |
Functional ≤ 10μM
|
|
LOX5-2-E |
Arachidonate 5-lipoxygenase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Functional ≤ 10μM
|
|
Z100741-2-O |
MC9 (Mast Cells) (cluster #2 Of 2), Other |
Other |
550 |
0.55 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
700 |
0.54 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
2400 |
0.49 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
2000 |
0.50 |
Functional ≤ 10μM
|
|
Z80419-1-O |
RBL-1 (Basophilic Leukemia Cells) (cluster #1 Of 2), Other |
Other |
100 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
2.54 |
-10.68 |
3 |
4 |
0 |
67 |
236.296 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.47 |
3.13 |
-43.84 |
2 |
4 |
-1 |
69 |
235.288 |
2 |
↓
|
|
|
|
Analogs
-
850
-
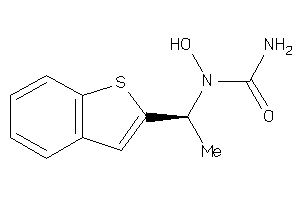
Draw
Identity
99%
90%
80%
70%
Vendors
And 32 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.51 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
830 |
0.53 |
Binding ≤ 10μM
|
|
LT4R1-1-E |
Leukotriene B4 Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
420 |
0.56 |
Binding ≤ 10μM
|
|
LKHA4-1-E |
Leukotriene A4 Hydrolase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
850 |
0.53 |
Functional ≤ 10μM
|
|
LOX5-2-E |
Arachidonate 5-lipoxygenase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Functional ≤ 10μM
|
|
Z100741-2-O |
MC9 (Mast Cells) (cluster #2 Of 2), Other |
Other |
550 |
0.55 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
700 |
0.54 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
2400 |
0.49 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
2000 |
0.50 |
Functional ≤ 10μM
|
|
Z80419-1-O |
RBL-1 (Basophilic Leukemia Cells) (cluster #1 Of 2), Other |
Other |
100 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
-3.7 |
-10.6 |
3 |
4 |
0 |
66 |
236.296 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.22 |
-3.5 |
-44.43 |
2 |
4 |
-1 |
70 |
235.288 |
3 |
↓
|
|