|
|
Analogs
-
13119547
-
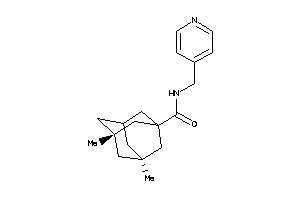
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7700 |
0.36 |
Binding ≤ 10μM
|
|
CP19A-2-E |
Cytochrome P450 19A1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1500 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
6.79 |
-7.41 |
1 |
3 |
0 |
42 |
270.376 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.80 |
7.24 |
-41.4 |
2 |
3 |
1 |
43 |
271.384 |
3 |
↓
|
|
|
|
Analogs
-
5356791
-
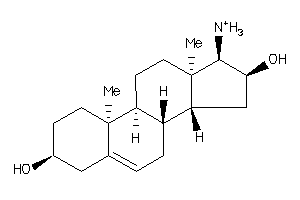
-
5356829
-
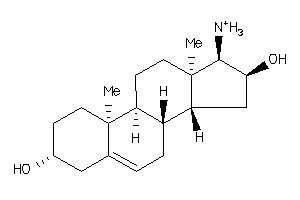
-
5359847
-
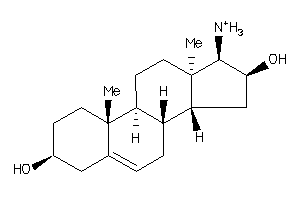
Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-aminoethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthr
17-(1-aminoethyl)-10,13-dimethyl…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9800 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
-2.73 |
-45.41 |
4 |
2 |
1 |
47 |
318.525 |
1 |
↓
|
|
|
|
Analogs
-
5356791
-

-
5356829
-

-
5359847
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-aminoethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthr
17-(1-aminoethyl)-10,13-dimethyl…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9800 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
-2.03 |
-42.99 |
4 |
2 |
1 |
47 |
318.525 |
1 |
↓
|
|
|
|
Analogs
-
5356791
-

-
5356829
-

-
5359847
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-aminoethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthr
17-(1-aminoethyl)-10,13-dimethyl…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9800 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
-2.78 |
-43.82 |
4 |
2 |
1 |
47 |
318.525 |
1 |
↓
|
|
|
|
Analogs
-
4451747
-
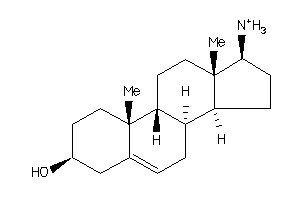
Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-aminoethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthr
17-(1-aminoethyl)-10,13-dimethyl…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9800 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
-2.35 |
-43.03 |
4 |
2 |
1 |
47 |
318.525 |
1 |
↓
|
|
|
|
Analogs
-
4532104
-
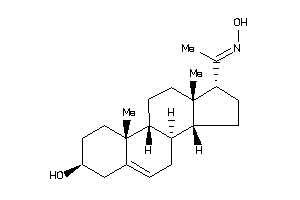
-
4532105
-
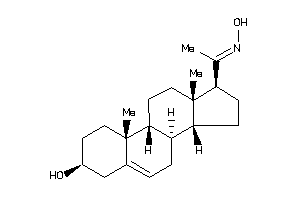
-
13759938
-
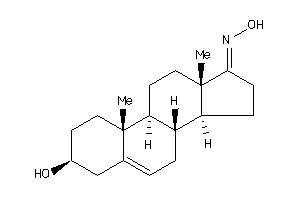
-
33938215
-
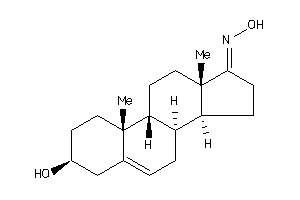
-
33938217
-
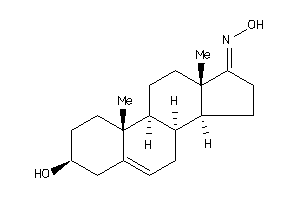
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.36 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1650 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
6.65 |
-4.75 |
2 |
3 |
0 |
53 |
331.5 |
1 |
↓
|
|
|
|
Analogs
-
13759938
-

-
33938215
-

-
33938217
-

-
4532102
-
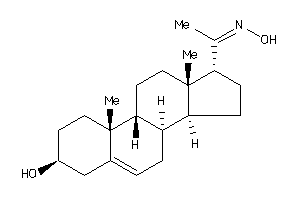
-
4532103
-
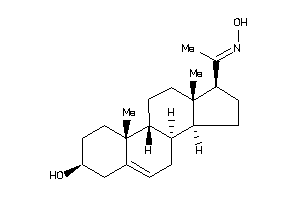
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.36 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1650 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
6.61 |
-4.44 |
2 |
3 |
0 |
53 |
331.5 |
1 |
↓
|
|
|
|
Analogs
-
3814414
-
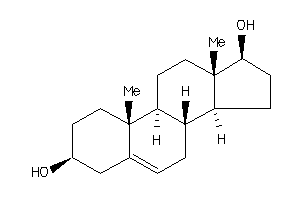
Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-hydroxyethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenant
17-(1-hydroxyethyl)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
-1.83 |
-2.97 |
2 |
2 |
0 |
40 |
318.501 |
1 |
↓
|
|
|
|
Analogs
-
3814414
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-hydroxyethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenant
17-(1-hydroxyethyl)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
-1.51 |
-2.75 |
2 |
2 |
0 |
40 |
318.501 |
1 |
↓
|
|
|
|
Analogs
-
3814414
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-hydroxyethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenant
17-(1-hydroxyethyl)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.41 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
-1.67 |
-2.64 |
2 |
2 |
0 |
40 |
318.501 |
1 |
↓
|
|
|
|
Analogs
-
3814414
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-hydroxyethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenant
17-(1-hydroxyethyl)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.41 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
-2.1 |
-2.77 |
2 |
2 |
0 |
40 |
318.501 |
1 |
↓
|
|
|
|
Analogs
-
103270
-
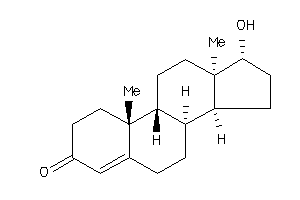
-
490793
-
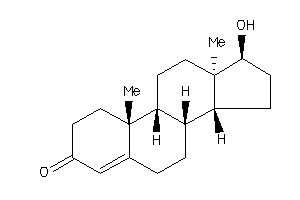
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9R,10R,13S,14S,17S)-17-(1-hydroxyethyl)-10,13-dimethyl-1,2,6,7,8,9,11,12,14,15,16,17-dodecahydro
(8R,9R,10R,13S,14S,17S)-17-(1-hy…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
490 |
0.38 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2840 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
8.26 |
-6.42 |
1 |
2 |
0 |
37 |
316.485 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
968234
-
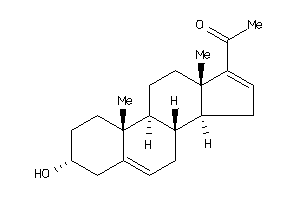
-
968235
-
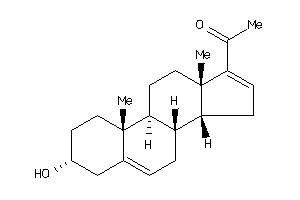
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15-decahydro-1H-cyclopenta[a]phenanthren-17-yl)etha
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
510 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
0.47 |
-8.51 |
1 |
2 |
0 |
37 |
314.469 |
1 |
↓
|
|
|
|
Analogs
-
968234
-

-
968235
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15-decahydro-1H-cyclopenta[a]phenanthren-17-yl)etha
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
510 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
1.01 |
-8.14 |
1 |
2 |
0 |
37 |
314.469 |
1 |
↓
|
|
|
|
Analogs
-
26392960
-
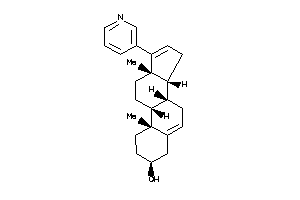
-
44830820
-
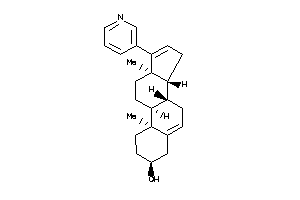
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
-0.59 |
-5.05 |
1 |
2 |
0 |
33 |
349.518 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
5359129
-
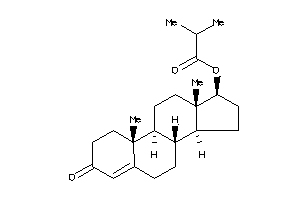
-
3875080
-
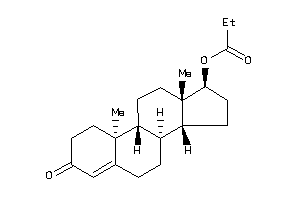
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
11.9 |
-10.74 |
0 |
3 |
0 |
43 |
344.495 |
3 |
↓
|
|
|
|
Analogs
-
3875080
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
11.95 |
-9.99 |
0 |
3 |
0 |
43 |
344.495 |
3 |
↓
|
|
|
|
Analogs
-
3875080
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
11.72 |
-10.5 |
0 |
3 |
0 |
43 |
344.495 |
3 |
↓
|
|
|
|
Analogs
-
10272298
-
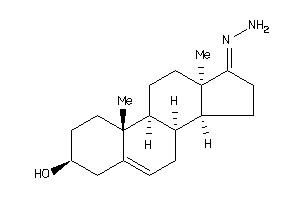
-
10272299
-
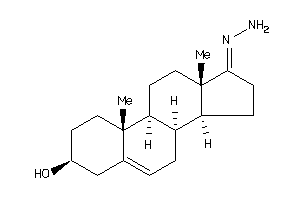
-
12719842
-
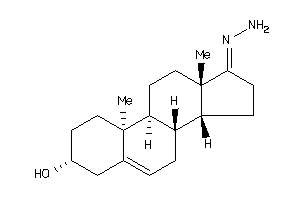
-
12719843
-
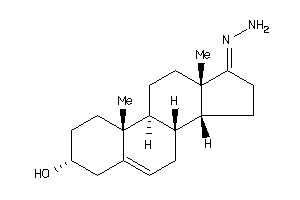
-
12719845
-
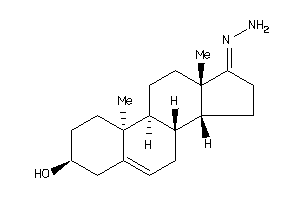
Draw
Identity
99%
90%
80%
70%
Popular Name:
17-hydrazono-10,13-dimethyl-1,2,3,4,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthren-3-ol
17-hydrazono-10,13-dimethyl-1,2,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6480 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
5.63 |
-4.4 |
3 |
3 |
0 |
59 |
302.462 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.40 |
5.53 |
-38.31 |
4 |
3 |
1 |
60 |
303.47 |
0 |
↓
|
|
|
|
Analogs
-
12719842
-

-
12719843
-

-
12719845
-

-
12719847
-
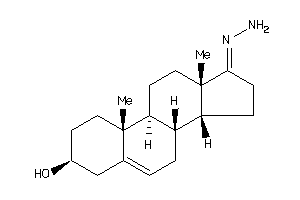
-
10272296
-
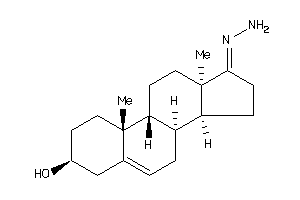
Draw
Identity
99%
90%
80%
70%
Popular Name:
17-hydrazono-10,13-dimethyl-1,2,3,4,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthren-3-ol
17-hydrazono-10,13-dimethyl-1,2,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6480 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
5.6 |
-4.55 |
3 |
3 |
0 |
59 |
302.462 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.40 |
5.48 |
-38.83 |
4 |
3 |
1 |
60 |
303.47 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,8R,9S,10R,13S,14S)-3-hydroxy-10,13-dimethyl-1,2,3,4,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a
(3S,8R,9S,10R,13S,14S)-3-hydroxy…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6480 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
5.5 |
-4.49 |
3 |
3 |
0 |
59 |
302.462 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,8R,9R,10R,13S,14S)-3-hydroxy-10,13-dimethyl-1,2,3,4,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a
(3S,8R,9R,10R,13S,14S)-3-hydroxy…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6480 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
5.59 |
-4.53 |
3 |
3 |
0 |
59 |
302.462 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.40 |
5.4 |
-41.22 |
4 |
3 |
1 |
60 |
303.47 |
0 |
↓
|
|
|
|
Analogs
-
4532103
-

-
4532104
-

-
4532105
-

-
13759938
-

-
33938215
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
6.6 |
-4.97 |
2 |
3 |
0 |
53 |
331.5 |
1 |
↓
|
|
|
|
Analogs
-
4532105
-

-
13759938
-

-
33938215
-

-
33938217
-

-
4532102
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
5.99 |
-5.35 |
2 |
3 |
0 |
53 |
331.5 |
1 |
↓
|
|
|
|
Analogs
-
753542
-
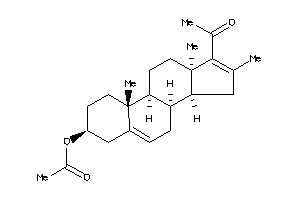
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1890 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
4.03 |
-10.32 |
0 |
3 |
0 |
43 |
356.506 |
3 |
↓
|
|
|
|
Analogs
-
753542
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1890 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
4.09 |
-9.65 |
0 |
3 |
0 |
43 |
356.506 |
3 |
↓
|
|
|
|
Analogs
-
753542
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1890 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
3.9 |
-10.61 |
0 |
3 |
0 |
43 |
356.506 |
3 |
↓
|
|
|
|
Analogs
-
753542
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(17-acetyl-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15-decahydro-1H-cyclopenta[a]phenanthren-3-yl)
(17-acetyl-10,13-dimethyl-2,3,4,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1890 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
3.89 |
-9.95 |
0 |
3 |
0 |
43 |
356.506 |
3 |
↓
|
|
|
|
Analogs
-
3982458
-
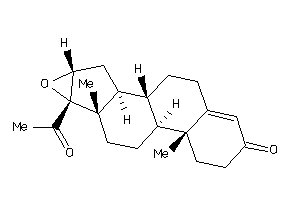
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
0.98 |
-9.45 |
0 |
3 |
0 |
46 |
328.452 |
1 |
↓
|
|
|
|
Analogs
-
4543816
-
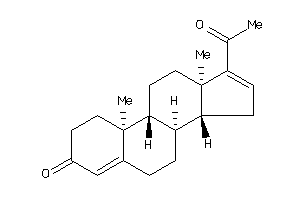
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10R,13S,14S)-17-acetyl-10,13-dimethyl-1,2,6,7,8,9,11,12,14,15-decahydrocyclopenta[a]phenanthr
(8S,9R,10R,13S,14S)-17-acetyl-10…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1770 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
10.54 |
-10.85 |
0 |
2 |
0 |
34 |
312.453 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
4369693
-
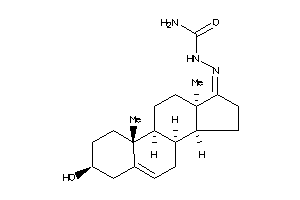
-
4369694
-
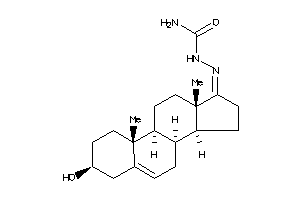
-
13467051
-
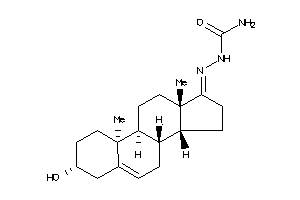
-
13467053
-
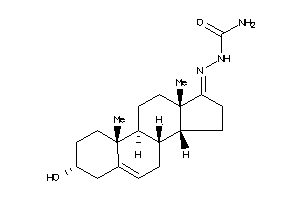
-
13467055
-
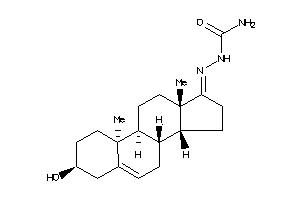
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yliden)hydrazine-1-carboxamide
2-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
178 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
3.82 |
-8.3 |
4 |
5 |
0 |
88 |
345.487 |
1 |
↓
|
|
|
|
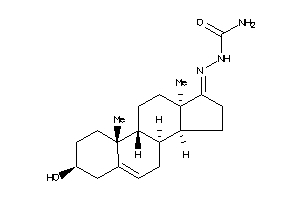
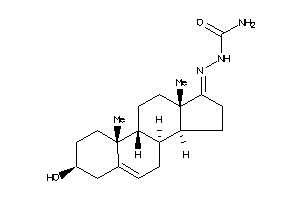
Analogs
-
4369691
-
-
4369692
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yliden)hydrazine-1-carboxamide
2-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
178 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
3.75 |
-8.54 |
4 |
5 |
0 |
88 |
345.487 |
1 |
↓
|
|
|
|
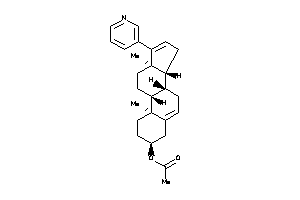
Analogs
-
44830832
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
18 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
13.26 |
-7.61 |
0 |
3 |
0 |
39 |
391.555 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.13 |
13.55 |
-35.52 |
1 |
3 |
1 |
40 |
392.563 |
3 |
↓
|
|
|
|
Analogs
-
44830832
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
18 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
13.93 |
-7.98 |
0 |
3 |
0 |
39 |
391.555 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.13 |
14.22 |
-38.07 |
1 |
3 |
1 |
40 |
392.563 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B2-1-E |
Cytochrome P450 11B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.41 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.67 |
1.15 |
-8.16 |
0 |
1 |
0 |
17 |
240.327 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B2-1-E |
Cytochrome P450 11B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.39 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.39 |
Binding ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
7420 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.26 |
1.85 |
-7.58 |
0 |
1 |
0 |
17 |
234.298 |
1 |
↓
|
|
|
|
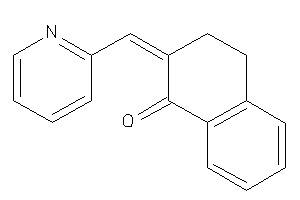
Analogs
-
5934749
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B2-1-E |
Cytochrome P450 11B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.39 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
1.44 |
-8.89 |
0 |
2 |
0 |
30 |
235.286 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.91 |
1.34 |
-37.8 |
1 |
2 |
1 |
31 |
236.294 |
1 |
↓
|
|