|
|
Analogs
-
26506281
-
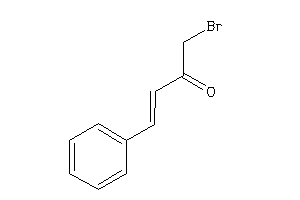
-
39128037
-
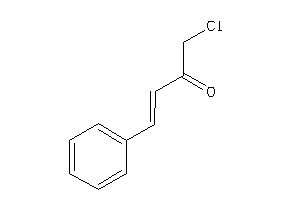
Draw
Identity
99%
90%
80%
70%
Vendors
And 129 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1A2-3-E |
Cytochrome P450 1A2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3700 |
0.42 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
10.65 |
-7.24 |
0 |
1 |
0 |
17 |
234.298 |
4 |
↓
|
|
|
|
Analogs
-
1550212
-
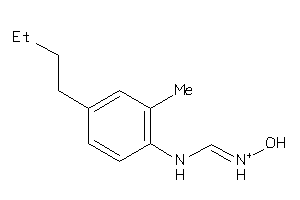
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
6.95 |
-6.04 |
2 |
3 |
0 |
45 |
206.289 |
5 |
↓
|
|
|
|
Analogs
-
38609341
-
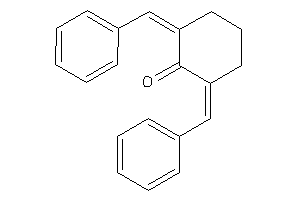
-
4582835
-
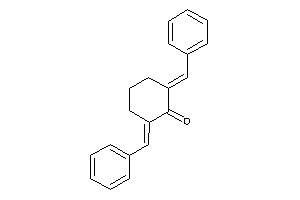
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1A2-3-E |
Cytochrome P450 1A2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.40 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.97 |
11.92 |
-6.62 |
0 |
1 |
0 |
17 |
274.363 |
2 |
↓
|
|
|
|
Analogs
-
3225
-
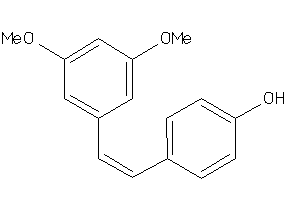
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AHR-1-E |
Aryl Hydrocarbon Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
261 |
0.46 |
Binding ≤ 10μM
|
|
ABL1-2-E |
Tyrosine-protein Kinase ABL (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Functional ≤ 10μM
|
|
BCR-2-E |
Breakpoint Cluster Region Protein (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Functional ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
830 |
0.43 |
ADME/T ≤ 10μM
|
|
CP1A2-3-E |
Cytochrome P450 1A2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6200 |
0.36 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
790 |
0.43 |
ADME/T ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
4000 |
0.38 |
Functional ≤ 10μM
|
|
Z80186-2-O |
K562 (Erythroleukemia Cells) (cluster #2 Of 11), Other |
Other |
10000 |
0.35 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1100 |
0.42 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
5700 |
0.37 |
Functional ≤ 10μM
|
|
Z81252-7-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #7 Of 11), Other |
Other |
1200 |
0.41 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
7.25 |
-7.23 |
0 |
3 |
0 |
28 |
270.328 |
5 |
↓
|
|
|
|
Analogs
-
3177729
-
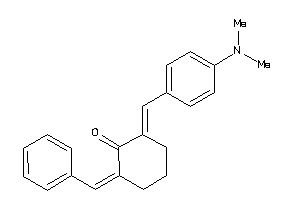
-
3194605
-
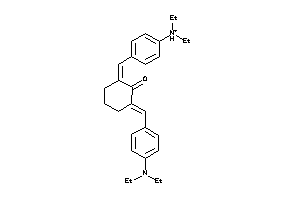
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1A2-3-E |
Cytochrome P450 1A2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.17 |
12.69 |
-8.61 |
0 |
3 |
0 |
24 |
360.501 |
4 |
↓
|
|
|
|
Analogs
-
7999104
-
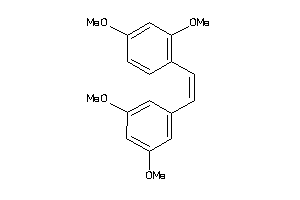
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.42 |
ADME/T ≤ 10μM
|
|
CP1A2-3-E |
Cytochrome P450 1A2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3100 |
0.35 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.52 |
ADME/T ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 3), Other |
Other |
8600 |
0.32 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
2.33 |
-8.72 |
0 |
4 |
0 |
36 |
300.354 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1A2-3-E |
Cytochrome P450 1A2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3400 |
0.38 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.49 |
11.69 |
-7.17 |
0 |
1 |
0 |
17 |
303.188 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1A2-3-E |
Cytochrome P450 1A2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.98 |
12.66 |
-8.72 |
0 |
1 |
0 |
17 |
370.292 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1A2-3-E |
Cytochrome P450 1A2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3400 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.28 |
12.96 |
-7.28 |
0 |
1 |
0 |
17 |
343.253 |
2 |
↓
|
|