|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
11.58 |
-40.63 |
1 |
4 |
1 |
31 |
288.415 |
9 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.24 |
12.76 |
-58.2 |
3 |
6 |
1 |
96 |
475.609 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
6.35 |
-85.33 |
5 |
4 |
2 |
63 |
329.831 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
5.92 |
-48.44 |
4 |
4 |
1 |
62 |
328.823 |
5 |
↓
|
|
|
|
Analogs
-
34606256
-
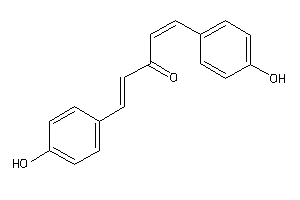
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
4.99 |
-10.19 |
2 |
3 |
0 |
58 |
266.296 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3900 |
0.30 |
Binding ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.29 |
ADME/T ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
6 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.29 |
8.78 |
-78.63 |
4 |
4 |
2 |
51 |
357.885 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.29 |
9.02 |
-33.48 |
2 |
4 |
0 |
52 |
355.869 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.48 |
7.13 |
-80.66 |
5 |
4 |
2 |
63 |
357.885 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.48 |
7.46 |
-32.55 |
3 |
4 |
0 |
65 |
355.869 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
940 |
0.53 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
6.82 |
-8.39 |
1 |
3 |
0 |
38 |
216.284 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.54 |
7.27 |
-31.87 |
2 |
3 |
1 |
39 |
217.292 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
40 |
0.80 |
ADME/T ≤ 10μM
|
|
CP2C9-3-E |
Cytochrome P450 2C9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
ADME/T ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5700 |
0.56 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.23 |
2.65 |
-48.73 |
3 |
3 |
1 |
54 |
175.211 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.23 |
2.93 |
-89.93 |
4 |
3 |
2 |
55 |
176.219 |
2 |
↓
|
|
|
|
Analogs
-
28564671
-
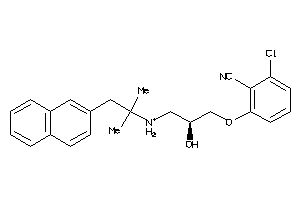
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.27 |
-0.48 |
-57.12 |
3 |
4 |
1 |
69 |
409.937 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.08 |
7.44 |
-94.67 |
7 |
7 |
2 |
113 |
432.572 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2200 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
5.78 |
-109.91 |
5 |
3 |
2 |
54 |
265.788 |
5 |
↓
|
|
|
|
Analogs
-
56556
-
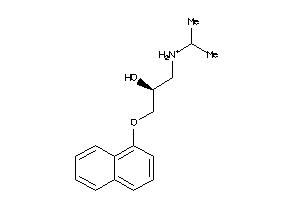
-
4071759
-
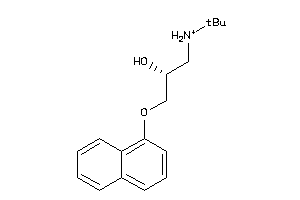
-
32102194
-
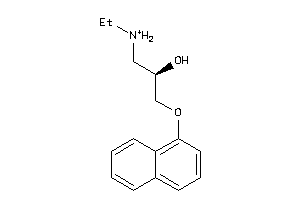
-
32102197
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 72 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
90 |
0.52 |
Binding ≤ 10μM
|
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.43 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9700 |
0.37 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.42 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.47 |
ADME/T ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
3162 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.45 |
-42.19 |
3 |
3 |
1 |
46 |
260.357 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.97 |
4.16 |
-6.9 |
2 |
3 |
0 |
41 |
259.349 |
6 |
↓
|
|
|
|
Analogs
-
4071759
-

-
32102194
-

-
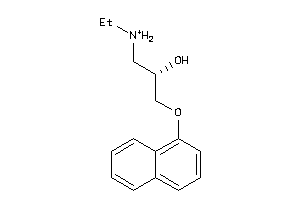
32102197
-

-
20240
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 69 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
55 |
0.53 |
Binding ≤ 10μM
|
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
90 |
0.52 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.48 |
Binding ≤ 10μM
|
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.48 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8100 |
0.38 |
Binding ≤ 10μM
|
|
5HT2B-3-E |
Serotonin 2b (5-HT2b) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.39 |
Binding ≤ 10μM
|
|
5HT2C-3-E |
Serotonin 2c (5-HT2c) Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.43 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9700 |
0.37 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
5HT1B-2-E |
Serotonin 1b (5-HT1b) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.40 |
Functional ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.43 |
ADME/T ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.42 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.48 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.47 |
ADME/T ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
3162 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.47 |
-42.1 |
3 |
3 |
1 |
46 |
260.357 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.97 |
4.28 |
-6.82 |
2 |
3 |
0 |
41 |
259.349 |
6 |
↓
|
|
|
|
Analogs
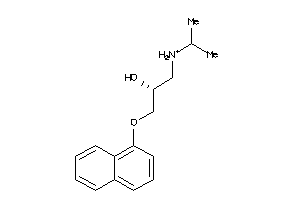
-
4212206
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.38 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
7.25 |
-52.17 |
4 |
5 |
1 |
86 |
364.469 |
8 |
↓
|
|
|
|
Analogs
-
1842330
-
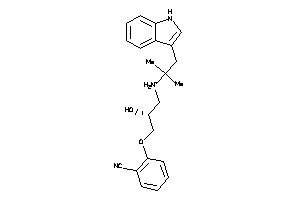
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.38 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
7.27 |
-52.11 |
4 |
5 |
1 |
86 |
364.469 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8000 |
0.51 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
2.75 |
-47.23 |
4 |
2 |
1 |
48 |
232.756 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.27 |
2.37 |
-5.47 |
3 |
2 |
0 |
46 |
231.748 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-2-E |
HERG (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
210 |
0.33 |
Binding ≤ 10μM
|
|
S22A1-1-E |
Solute Carrier Family 22 Member 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9940 |
0.25 |
Binding ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.31 |
Binding ≤ 10μM |
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Functional ≤ 10μM
|
|
ADO-1-E |
Aldehyde Oxidase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6400 |
0.26 |
ADME/T ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.26 |
ADME/T ≤ 10μM
|
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
5890 |
0.26 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
600 |
0.31 |
Functional ≤ 10μM
|
|
Z50460-4-O |
Leishmania Major (cluster #4 Of 4), Other |
Other |
1500 |
0.29 |
Functional ≤ 10μM
|
|
Z80064-8-O |
CCRF-CEM (T-cell Leukemia) (cluster #8 Of 9), Other |
Other |
139 |
0.34 |
Functional ≤ 10μM
|
|
Z80084-1-O |
CEM-VLB (Vinblastine-resistant T-cell Leukemia Cells) (cluster #1 Of 2), Other |
Other |
520 |
0.31 |
Functional ≤ 10μM
|
|
Z80085-1-O |
CEM-VM1 (Etoposide-resistant T-cell Leukemia Cells) (cluster #1 Of 2), Other |
Other |
1910 |
0.29 |
Functional ≤ 10μM
|
|
Z80089-1-O |
CHO-AA8 (cluster #1 Of 2), Other |
Other |
1 |
0.45 |
Functional ≤ 10μM
|
|
Z80115-1-O |
DC3F (cluster #1 Of 1), Other |
Other |
4 |
0.42 |
Functional ≤ 10μM
|
|
Z80117-2-O |
DC3F/AD-II (cluster #2 Of 2), Other |
Other |
47 |
0.37 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
120 |
0.35 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
6 |
0.41 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
559 |
0.31 |
Functional ≤ 10μM |
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
23 |
0.38 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
50 |
0.37 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
75 |
0.36 |
Functional ≤ 10μM
|
|
Z80227-2-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
1165 |
0.30 |
Functional ≤ 10μM
|
|
Z80244-7-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #7 Of 7), Other |
Other |
490 |
0.32 |
Functional ≤ 10μM
|
|
Z80322-1-O |
NCI-H358 (Lung Carcinama Cells) (cluster #1 Of 2), Other |
Other |
150 |
0.34 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
890 |
0.30 |
Functional ≤ 10μM
|
|
Z80475-3-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #3 Of 3), Other |
Other |
60 |
0.36 |
Functional ≤ 10μM
|
|
Z80601-1-O |
XRS6 (cluster #1 Of 1), Other |
Other |
240 |
0.33 |
Functional ≤ 10μM
|
|
Z80608-4-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #4 Of 4), Other |
Other |
180 |
0.34 |
Functional ≤ 10μM
|
|
Z80612-1-O |
2008 (Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
4310 |
0.27 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
3520 |
0.27 |
Functional ≤ 10μM
|
|
Z80712-7-O |
T47D (Breast Carcinoma Cells) (cluster #7 Of 7), Other |
Other |
220 |
0.33 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
1485 |
0.29 |
Functional ≤ 10μM |
|
Z80846-2-O |
F460pv8/eto Cell Line (cluster #2 Of 2), Other |
Other |
40 |
0.37 |
Functional ≤ 10μM
|
|
Z80889-2-O |
NCI-H322M (Non-small Cell Lung Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
210 |
0.33 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
700 |
0.31 |
Functional ≤ 10μM
|
|
Z81024-8-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #8 Of 8), Other |
Other |
60 |
0.36 |
Functional ≤ 10μM
|
|
Z81034-6-O |
A2780 (Ovarian Carcinoma Cells) (cluster #6 Of 10), Other |
Other |
100 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-6-O |
Jurkat (Acute Leukemic T-cells) (cluster #6 Of 10), Other |
Other |
80 |
0.35 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z81164-1-O |
LL Cell Line (cluster #1 Of 2), Other |
Other |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z81167-1-O |
LLTC Cell Line (cluster #1 Of 1), Other |
Other |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
901 |
0.30 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
9500 |
0.25 |
Functional ≤ 10μM
|
|
Z81252-10-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #10 Of 11), Other |
Other |
140 |
0.34 |
Functional ≤ 10μM
|
|
Z81280-1-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
1010 |
0.30 |
Functional ≤ 10μM
|
|
Z81284-2-O |
NCI-H647 (cluster #2 Of 2), Other |
Other |
70 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.91 |
6.33 |
-44.52 |
2 |
6 |
0 |
84 |
393.468 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.91 |
5.83 |
-16.62 |
2 |
6 |
0 |
80 |
393.468 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.91 |
5.94 |
-47.33 |
1 |
6 |
-1 |
82 |
392.46 |
5 |
↓
|
|
|
|
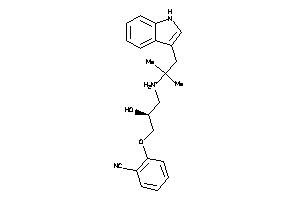
Analogs
-
527387
-
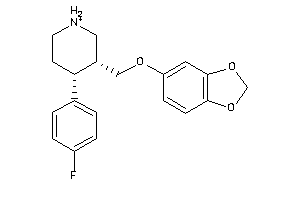
-
527385
-
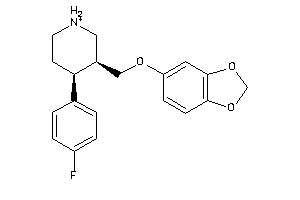
Draw
Identity
99%
90%
80%
70%
Vendors
And 61 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NORA-1-B |
Quinolone Resistance Protein NorA (cluster #1 Of 1), Bacterial |
Bacteria |
7000 |
0.30 |
Binding ≤ 10μM |
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6310 |
0.30 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6310 |
0.30 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6310 |
0.30 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
59 |
0.42 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.36 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.41 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.37 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.48 |
Functional ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4850 |
0.31 |
ADME/T ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
5320 |
0.31 |
Binding ≤ 10μM |
|
Z50185-3-O |
Staphylococcus Aureus (cluster #3 Of 4), Other |
Other |
10000 |
0.29 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.30 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
7620 |
0.30 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
7.73 |
-53.07 |
2 |
4 |
1 |
44 |
330.379 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHB1-1-E |
Estradiol 17-beta-dehydrogenase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
46 |
0.51 |
Binding ≤ 10μM
|
|
DHB2-1-E |
Estradiol 17-beta-dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1971 |
0.40 |
Binding ≤ 10μM
|
|
CP2CJ-1-E |
Cytochrome P450 2C19 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
940 |
0.42 |
ADME/T ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1500 |
0.41 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.44 |
ADME/T ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
426 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.30 |
5.98 |
-8 |
2 |
2 |
0 |
40 |
282.364 |
2 |
↓
|
|
|
|
Analogs
-
32108748
-
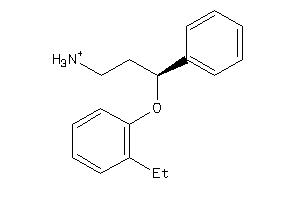
-
39298020
-
-
39298021
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2100 |
0.42 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4383 |
0.39 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4186 |
0.40 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.62 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.61 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.61 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3100 |
0.41 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
77 |
0.52 |
Binding ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.42 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
1585 |
0.43 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
4 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
8.89 |
-40.02 |
2 |
2 |
1 |
26 |
256.369 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.29 |
8.56 |
-80.64 |
4 |
4 |
2 |
51 |
357.885 |
6 |
↓
|
|