|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.57 |
15.05 |
-43.07 |
1 |
4 |
1 |
28 |
461.457 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.57 |
12.6 |
-12.35 |
0 |
4 |
0 |
27 |
460.449 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.35 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.25 |
Functional ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
750 |
0.28 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.26 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.57 |
14.73 |
-27.78 |
1 |
4 |
1 |
28 |
461.457 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.57 |
13.88 |
-10.16 |
0 |
4 |
0 |
27 |
460.449 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1930 |
0.38 |
Binding ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.34 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
8.71 |
-45.39 |
2 |
2 |
1 |
20 |
322.259 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.17 |
7.3 |
-2.76 |
1 |
2 |
0 |
15 |
321.251 |
3 |
↓
|
|
|
|
Analogs
-
3973106
-
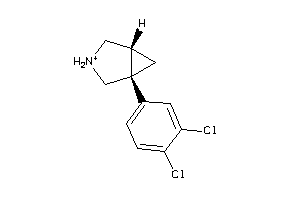
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.52 |
6.98 |
-45.38 |
2 |
1 |
1 |
17 |
229.13 |
1 |
↓
|
|
|
|
Analogs
-
3189
-
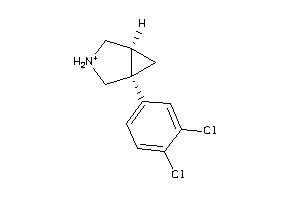
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.50 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
88 |
0.71 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
430 |
0.64 |
Binding ≤ 10μM
|
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
72 |
0.71 |
Binding ≤ 10μM
|
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
794 |
0.61 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
38 |
0.74 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
96 |
0.70 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.76 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
66 |
0.72 |
Binding ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.52 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.52 |
-0.86 |
-46.72 |
2 |
1 |
1 |
16 |
229.13 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
63 |
0.42 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
50 |
0.43 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
63 |
0.42 |
Binding ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.43 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
12.68 |
-41.47 |
1 |
3 |
1 |
25 |
323.46 |
6 |
↓
|
|
|
|
Analogs
-
56446
-
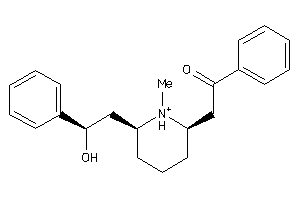
-
156831
-
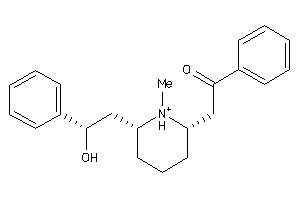
-
156931
-
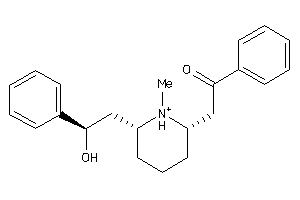
-
519487
-
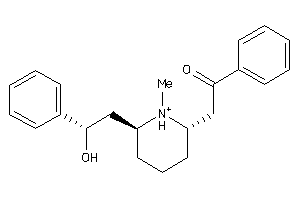
-
1081316
-
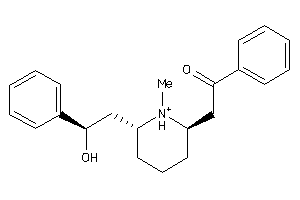
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA2-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA5-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA6-3-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6260 |
0.29 |
Binding ≤ 10μM
|
|
ACHA9-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB2-5-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB3-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB4-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2760 |
0.31 |
Binding ≤ 10μM
|
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6260 |
0.29 |
Binding ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.39 |
ADME/T ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
4 |
0.47 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
40 |
0.41 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
10.48 |
-45.25 |
2 |
3 |
1 |
42 |
338.471 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.73 |
8.25 |
-8.89 |
1 |
3 |
0 |
41 |
337.463 |
6 |
↓
|
|
|
|
Analogs
-
519487
-

-
1624
-
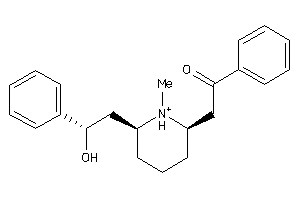
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6260 |
0.29 |
Binding ≤ 10μM
|
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2760 |
0.31 |
Binding ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.39 |
ADME/T ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
4 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
10.67 |
-44.31 |
2 |
3 |
1 |
42 |
338.471 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.73 |
8.5 |
-7.31 |
1 |
3 |
0 |
41 |
337.463 |
6 |
↓
|
|
|
|
Analogs
-
17950
-
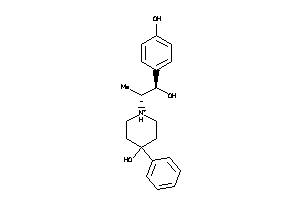
-
13742659
-
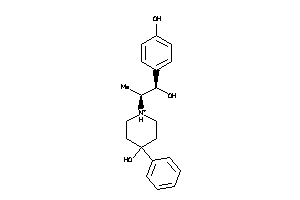
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.29 |
Binding ≤ 10μM
|
|
NMD3A-3-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
27 |
0.44 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
27 |
0.44 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.48 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.48 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.48 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.48 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.48 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
74 |
0.42 |
Functional ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.42 |
Functional ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4250 |
0.31 |
ADME/T ≤ 10μM
|
|
Z104297-2-O |
Glutamate NMDA Receptor; GRIN1/GRIN2B (cluster #2 Of 3), Other |
Other |
30 |
0.44 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
5700 |
0.31 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 3), Other |
Other |
11 |
0.46 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
11 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
-3.61 |
-37.03 |
4 |
4 |
1 |
65 |
328.432 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.68 |
9.21 |
-47.6 |
2 |
5 |
1 |
55 |
356.49 |
4 |
↓
|
|
|
|
Analogs
-
4212951
-
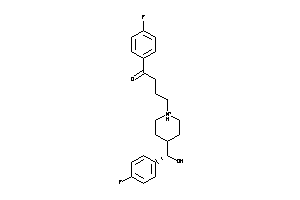
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1600 |
0.30 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
1.47 |
-49.69 |
2 |
3 |
1 |
41 |
374.451 |
7 |
↓
|
|
|
|
Analogs
-
1579717
-
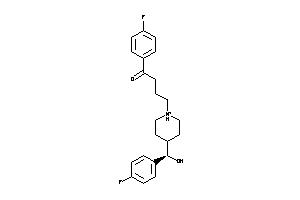
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1600 |
0.30 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
1.61 |
-49.79 |
2 |
3 |
1 |
41 |
374.451 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2100 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
6.68 |
-39.38 |
2 |
2 |
1 |
20 |
306.282 |
5 |
↓
|
|