|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1415 |
0.24 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Binding ≤ 10μM
|
|
CCR1-1-E |
C-C Chemokine Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
CCR2-1-E |
C-C Chemokine Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
CCR3-1-E |
C-C Chemokine Receptor Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.33 |
Binding ≤ 10μM
|
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Binding ≤ 10μM
|
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
611 |
0.26 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
611 |
0.26 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
718 |
0.25 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2350 |
0.23 |
Binding ≤ 10μM
|
|
CCR3-1-E |
C-C Chemokine Receptor Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.33 |
Functional ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.27 |
ADME/T ≤ 10μM
|
|
DRD2-17-E |
Dopamine D2 Receptor (cluster #17 Of 24), Eukaryotic |
Eukaryotes |
321 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.40 |
14.84 |
-55.42 |
3 |
6 |
1 |
76 |
487.665 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.73 |
3.67 |
-17.66 |
2 |
8 |
0 |
112 |
397.431 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.73 |
3.61 |
-19.86 |
2 |
8 |
0 |
112 |
397.431 |
6 |
↓
|
|
|
|
Analogs
-
4827283
-
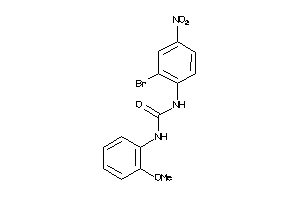
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
4.46 |
-9.29 |
3 |
7 |
0 |
107 |
352.144 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.37 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.55 |
Binding ≤ 10μM
|
|
Z50587-3-O |
Homo Sapiens (cluster #3 Of 9), Other |
Other |
1 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
-2.94 |
-8.61 |
3 |
5 |
0 |
85 |
411.053 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.45 |
Binding ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
339 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
-2.52 |
-11.33 |
3 |
7 |
0 |
107 |
273.248 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.51 |
Binding ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
10 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
4.18 |
-37.84 |
2 |
7 |
-1 |
105 |
356.163 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.91 |
4.02 |
-10.58 |
3 |
7 |
0 |
106 |
357.171 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.30 |
Binding ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
170 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.46 |
-1.85 |
-40.05 |
2 |
4 |
1 |
42 |
506.548 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
99 |
0.34 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.73 |
3.67 |
-17.19 |
2 |
8 |
0 |
112 |
397.431 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.73 |
3.61 |
-20.41 |
2 |
8 |
0 |
112 |
397.431 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
950 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
-2.7 |
-12.4 |
3 |
7 |
0 |
107 |
307.693 |
3 |
↓
|
|