|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 43 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.34 |
Binding ≤ 10μM
|
|
DHI2-1-E |
11-beta-hydroxysteroid Dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
257 |
0.27 |
Binding ≤ 10μM
|
|
KPCL-1-E |
Protein Kinase C Eta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
250 |
0.27 |
Binding ≤ 10μM |
|
PTN11-2-E |
Protein-tyrosine Phosphatase 2C (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9600 |
0.21 |
Binding ≤ 10μM
|
|
Z80418-3-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #3 Of 9), Other |
Other |
1300 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
11.48 |
-62.34 |
1 |
4 |
-1 |
77 |
469.686 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
5.62 |
11.49 |
-61.44 |
1 |
4 |
-1 |
77 |
469.686 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.62 |
9.52 |
-14.08 |
2 |
4 |
0 |
75 |
470.694 |
1 |
↓
|
|
|
|
Analogs
-
3977823
-
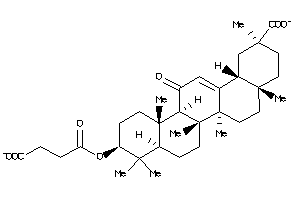
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.23 |
Binding ≤ 10μM
|
|
DHI2-1-E |
11-beta-hydroxysteroid Dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
82 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.60 |
17.01 |
-110.52 |
0 |
7 |
-2 |
124 |
568.751 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.60 |
15.05 |
-56.75 |
1 |
7 |
-1 |
121 |
569.759 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHB2-2-E |
Estradiol 17-beta-dehydrogenase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3780 |
0.21 |
Binding ≤ 10μM
|
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.23 |
Binding ≤ 10μM
|
|
DHI2-1-E |
11-beta-hydroxysteroid Dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
750 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.33 |
15.57 |
-67.71 |
0 |
5 |
-1 |
84 |
511.723 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.33 |
13.6 |
-17.98 |
1 |
5 |
0 |
81 |
512.731 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
4-(3-acetoxy-4,4,10,13,14-pentamethyl-7,11-dioxo-2,3,5,6,12,15,16,17-octahydro-1H-cyclopenta[a]phena
4-(3-acetoxy-4,4,10,13,14-pentam…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
410 |
0.26 |
Binding ≤ 10μM
|
|
DHI2-1-E |
11-beta-hydroxysteroid Dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3950 |
0.22 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2090 |
0.23 |
Functional ≤ 10μM
|
|
Z80936-2-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #2 Of 4), Other |
Other |
2090 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
4.91 |
-53.26 |
0 |
6 |
-1 |
100 |
485.641 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,4aS,6aR,6aS,6bR,8aR,10S,12aS,14bR)-10-amino-2,4a,6a,6b,9,9,12a-heptamethyl-13-oxo-3,4,5,6,6a,7,8
(2S,4aS,6aR,6aS,6bR,8aR,10S,12aS…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
172 |
0.28 |
Binding ≤ 10μM
|
|
DHI2-1-E |
11-beta-hydroxysteroid Dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6040 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
12.05 |
-96.16 |
3 |
4 |
0 |
85 |
469.71 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.13 |
10.09 |
-53.35 |
4 |
4 |
1 |
82 |
470.718 |
1 |
↓
|
|
|
|
|