|
|
|
|
|
Analogs
-
34590727
-
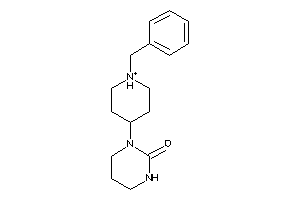
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP2-2-E |
Dipeptidyl Peptidase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
9.62 |
-45.96 |
2 |
4 |
1 |
37 |
302.442 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP2-2-E |
Dipeptidyl Peptidase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
277 |
0.92 |
Binding ≤ 10μM
|
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7880 |
0.71 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.57 |
2.1 |
-43.12 |
3 |
2 |
1 |
31 |
177.318 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
13522765
-
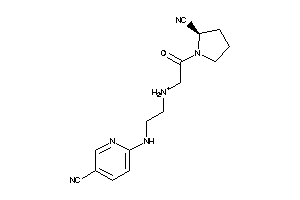
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP2-2-E |
Dipeptidyl Peptidase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6600 |
0.33 |
Binding ≤ 10μM
|
|
DPP4-1-E |
Dipeptidyl Peptidase IV (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4573 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.38 |
4.95 |
-65.32 |
3 |
7 |
1 |
109 |
299.358 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.38 |
3.53 |
-18.36 |
2 |
7 |
0 |
105 |
298.35 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP2-2-E |
Dipeptidyl Peptidase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1430 |
0.82 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.54 |
2.52 |
-40.96 |
3 |
2 |
1 |
31 |
159.278 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP2-2-E |
Dipeptidyl Peptidase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.60 |
Binding ≤ 10μM
|
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
5.67 |
-59.06 |
4 |
4 |
1 |
63 |
310.849 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.57 |
4.65 |
-47.94 |
4 |
4 |
1 |
60 |
310.849 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.57 |
4.35 |
-7.77 |
3 |
4 |
0 |
58 |
309.841 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPB-1-E |
Aminopeptidase B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.54 |
Binding ≤ 10μM |
|
AMPE-2-E |
Aminopeptidase A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
380 |
0.43 |
Binding ≤ 10μM
|
|
AMPN-2-E |
Aminopeptidase N (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.54 |
Binding ≤ 10μM
|
|
AMPN-2-E |
Aminopeptidase N (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
380 |
0.43 |
Binding ≤ 10μM
|
|
DPP2-2-E |
Dipeptidyl Peptidase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.60 |
Binding ≤ 10μM
|
|
DPP3-1-E |
Dipeptidyl Peptidase III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.60 |
Binding ≤ 10μM
|
|
DPP3-1-E |
Dipeptidyl Peptidase III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.58 |
Binding ≤ 10μM
|
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.60 |
Binding ≤ 10μM
|
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.58 |
Binding ≤ 10μM
|
|
DPP6-1-E |
Dipeptidyl Peptidase VI (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.60 |
Binding ≤ 10μM
|
|
ECE1-1-E |
Endothelin-converting Enzyme 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.54 |
Binding ≤ 10μM
|
|
LKHA4-1-E |
Leukotriene A4 Hydrolase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.55 |
Binding ≤ 10μM
|
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.58 |
Binding ≤ 10μM
|
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.54 |
Binding ≤ 10μM
|
|
THER-1-B |
Thermolysin (cluster #1 Of 3), Bacterial |
Bacteria |
3 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.23 |
3.01 |
-61.22 |
3 |
7 |
-1 |
119 |
293.299 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.23 |
4.26 |
-116.92 |
2 |
7 |
-2 |
121 |
292.291 |
7 |
↓
|
|