|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1720 |
0.50 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2430 |
0.49 |
Binding ≤ 10μM
|
|
Z80224-12-O |
MCF7 (Breast Carcinoma Cells) (cluster #12 Of 14), Other |
Other |
970 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
4.74 |
-15.97 |
1 |
4 |
0 |
54 |
298.165 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
1.96 |
5.69 |
-7.92 |
1 |
4 |
0 |
54 |
298.165 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
90 |
0.58 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
420 |
0.53 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
60 |
0.59 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
3.19 |
-8.36 |
2 |
5 |
0 |
74 |
314.164 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.88 |
3.23 |
-9.1 |
2 |
5 |
0 |
74 |
314.164 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
2.41 |
2.24 |
-14.82 |
2 |
5 |
0 |
74 |
314.164 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1460 |
0.45 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4510 |
0.42 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1560 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
2.68 |
-16.14 |
2 |
5 |
0 |
74 |
304.158 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.14 |
4.43 |
-41.6 |
1 |
5 |
-1 |
77 |
303.15 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.68 |
3.46 |
-50.77 |
1 |
5 |
-1 |
77 |
303.15 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
5124238
-
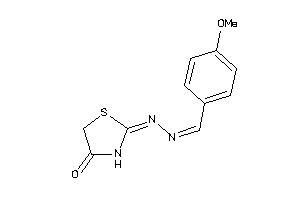
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5780 |
0.43 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9170 |
0.41 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
3160 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
4.4 |
-10.31 |
1 |
5 |
0 |
63 |
249.295 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
1.74 |
3.43 |
-17.55 |
1 |
5 |
0 |
63 |
249.295 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
1.21 |
4.37 |
-9.47 |
1 |
5 |
0 |
63 |
249.295 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6820 |
0.40 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8460 |
0.39 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
2770 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.11 |
4.99 |
-13.85 |
1 |
7 |
0 |
100 |
264.266 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
1.11 |
5 |
-15.03 |
1 |
7 |
0 |
100 |
264.266 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
1.64 |
4.83 |
-20.79 |
1 |
7 |
0 |
100 |
264.266 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2180 |
0.50 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3890 |
0.47 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1580 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.85 |
4.18 |
-16.63 |
1 |
4 |
0 |
54 |
237.259 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
1.31 |
5.16 |
-9.11 |
1 |
4 |
0 |
54 |
237.259 |
2 |
↓
|
|
|
|
Analogs
-
38194752
-
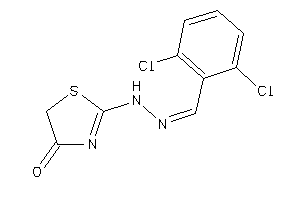
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1940 |
0.50 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2750 |
0.49 |
Binding ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
1820 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
5.58 |
-7.97 |
1 |
4 |
0 |
54 |
253.714 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.83 |
5.61 |
-8.68 |
1 |
4 |
0 |
54 |
253.714 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
2.36 |
4.64 |
-15.98 |
1 |
4 |
0 |
54 |
253.714 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
17045393
-
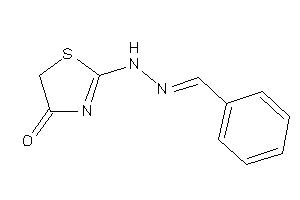
-
13633783
-
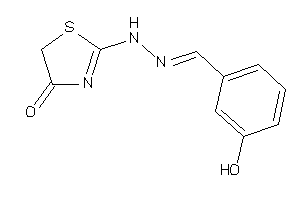
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4710 |
0.47 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6930 |
0.45 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1670 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
1.27 |
-17.58 |
2 |
5 |
0 |
74 |
235.268 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
0.67 |
2.25 |
-10.41 |
2 |
5 |
0 |
74 |
235.268 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.67 |
3 |
-47.3 |
1 |
5 |
-1 |
77 |
234.26 |
2 |
↓
|
|