|
|
Analogs
-
3830196
-
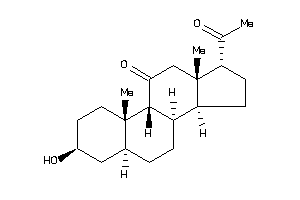
-
3830197
-
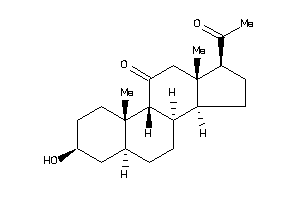
-
3830198
-
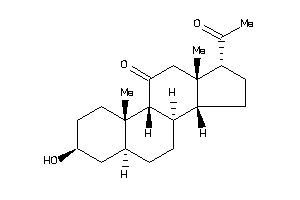
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA2-4-E |
GABA Receptor Alpha-2 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
2000 |
0.33 |
Binding ≤ 10μM
|
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
2000 |
0.33 |
Binding ≤ 10μM
|
|
GBRG2-3-E |
GABA Receptor Gamma-2 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
2000 |
0.33 |
Binding ≤ 10μM
|
|
Z104301-2-O |
GABA-A Receptor; Anion Channel (cluster #2 Of 8), Other |
Other |
300 |
0.38 |
Binding ≤ 10μM
|
|
Z50573-2-O |
Xenopus Laevis (cluster #2 Of 2), Other |
Other |
5480 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
7.55 |
-8.46 |
1 |
3 |
0 |
54 |
332.484 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
6093626
-
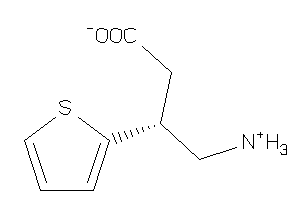
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-2-E |
GABA-B Receptor 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9720 |
0.58 |
Binding ≤ 10μM
|
|
GABR2-2-E |
GABA-B Receptor 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9720 |
0.58 |
Binding ≤ 10μM
|
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
9720 |
0.58 |
Binding ≤ 10μM
|
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
9720 |
0.58 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
9720 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.67 |
2.69 |
-50.54 |
3 |
3 |
0 |
68 |
185.248 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.22 |
-1.97 |
-67.58 |
2 |
4 |
0 |
65 |
159.185 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.22 |
1.79 |
-68.69 |
2 |
4 |
0 |
66 |
159.185 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.22 |
0.46 |
-50.59 |
1 |
4 |
-1 |
61 |
158.177 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.22 |
1.68 |
-70.35 |
2 |
4 |
0 |
66 |
159.185 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.22 |
0.34 |
-48.74 |
1 |
4 |
-1 |
61 |
158.177 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
9000 |
0.64 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.22 |
1.81 |
-68.69 |
2 |
4 |
0 |
66 |
159.185 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.22 |
0.47 |
-48.82 |
1 |
4 |
-1 |
61 |
158.177 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
-1.63 |
-64.6 |
2 |
4 |
0 |
65 |
173.212 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
2.45 |
-68.71 |
2 |
4 |
0 |
66 |
173.212 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.31 |
1.12 |
-50.35 |
1 |
4 |
-1 |
61 |
172.204 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
2.46 |
-68.74 |
2 |
4 |
0 |
66 |
173.212 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.31 |
1.26 |
-48.68 |
1 |
4 |
-1 |
61 |
172.204 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
2.6 |
-67.02 |
2 |
4 |
0 |
66 |
173.212 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.31 |
1.4 |
-48.68 |
1 |
4 |
-1 |
61 |
172.204 |
3 |
↓
|
|
|
|
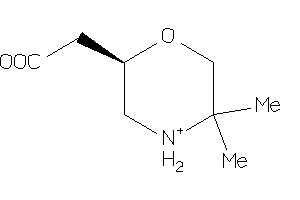
Analogs
-
2546079
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM |
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM |
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM |
|
Z50512-2-O |
Cavia Porcellus (cluster #2 Of 7), Other |
Other |
4600 |
0.62 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.26 |
3.23 |
-60.75 |
2 |
4 |
0 |
66 |
173.212 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.26 |
2.03 |
-48.26 |
1 |
4 |
-1 |
61 |
172.204 |
2 |
↓
|
|
|
|
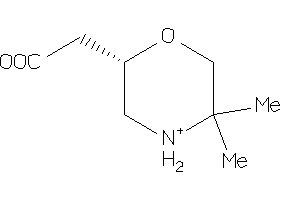
Analogs
-
6164
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM |
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM |
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM |
|
Z50512-2-O |
Cavia Porcellus (cluster #2 Of 7), Other |
Other |
4600 |
0.62 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.26 |
3.23 |
-60.76 |
2 |
4 |
0 |
66 |
173.212 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.26 |
2.02 |
-47.01 |
1 |
4 |
-1 |
61 |
172.204 |
2 |
↓
|
|